Engineering Insect Symbionts
Many insects have more consequential associations with bacteria than we have with the human microbiome. Recently, we have begun to genetically engineer these bacteria to study these symbioses and as a way of controlling their insect hosts. These projects are highly collaborative and have also involved researchers from the
Moran Lab, the
Ellington Lab, and other labs.
 Funding:
Funding: DARPA BRICS, DARPA Insect Allies, ARO MURI
Honey Bees: Protecting Pollinator Health
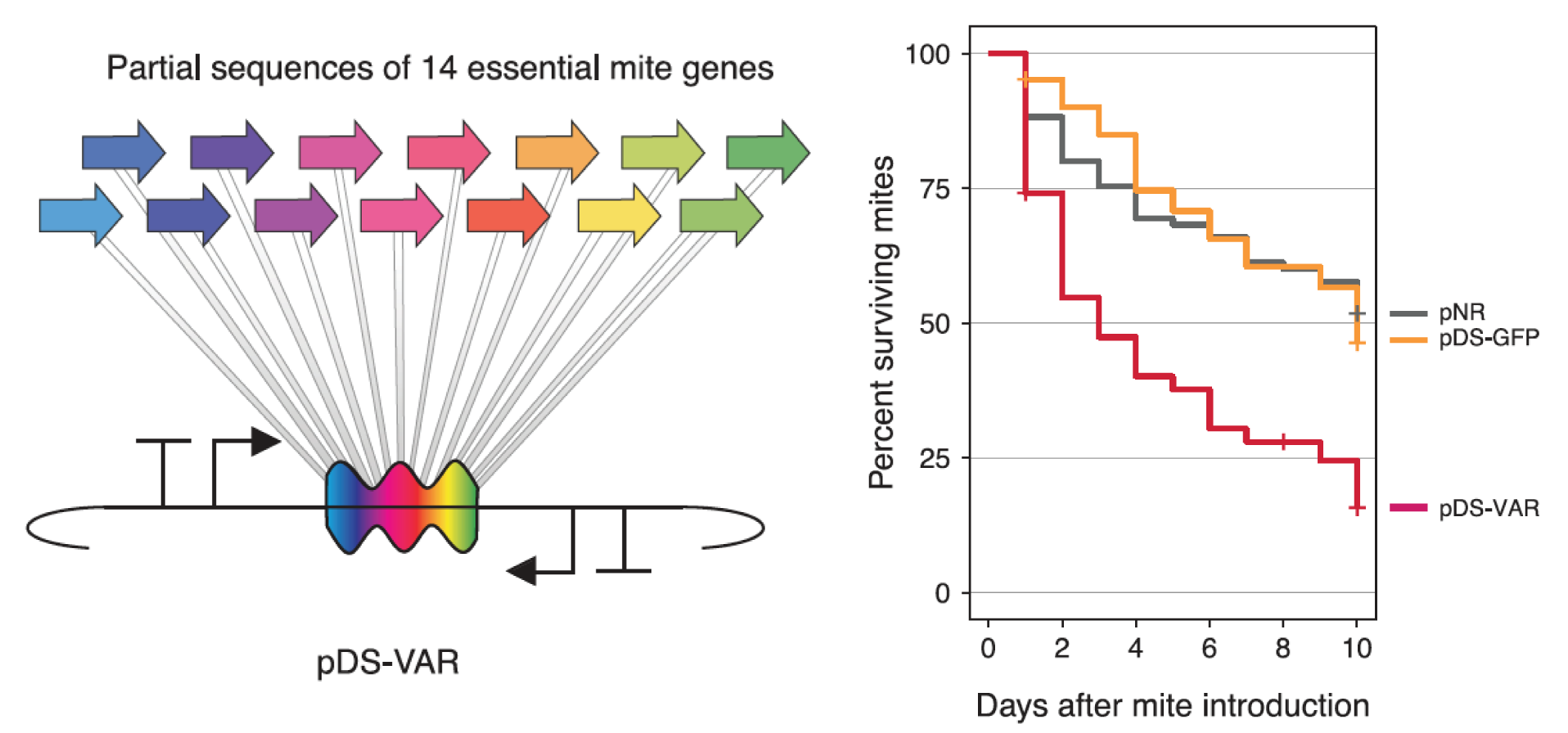
Engineered symbionts kill honey bee parasites
Bees are ecologically and economically important pollinators. We created a bee microbiome toolkit (BTK) for engineering bacteria that colonize the guts of honey bees (
Apis mellifera) and bumble bees (
Bombus species). We recently showed that we can engineer the bacterium
Snodgrassella alvi to implement effective symbiont-mediated RNAi. This system can be used for the targeted knockdown of bee genes to study their functions. For example, we have used it to alter honey bee feeding behavior. We have also shown that these engineered symbionts can be used to protect honey bees from viral pathogens and arthropod parasites that imperil the health of hives.
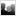 Researchers: Sean Leonard
Researchers: Sean Leonard,
Peng Geng
 News: Bacteria Engineered to Protect Bees from Pests and Pathogens
News: Bacteria Engineered to Protect Bees from Pests and Pathogens
 Representative Publications
Representative Publications Aphids: Understanding the Evolution of an Endosymbiont
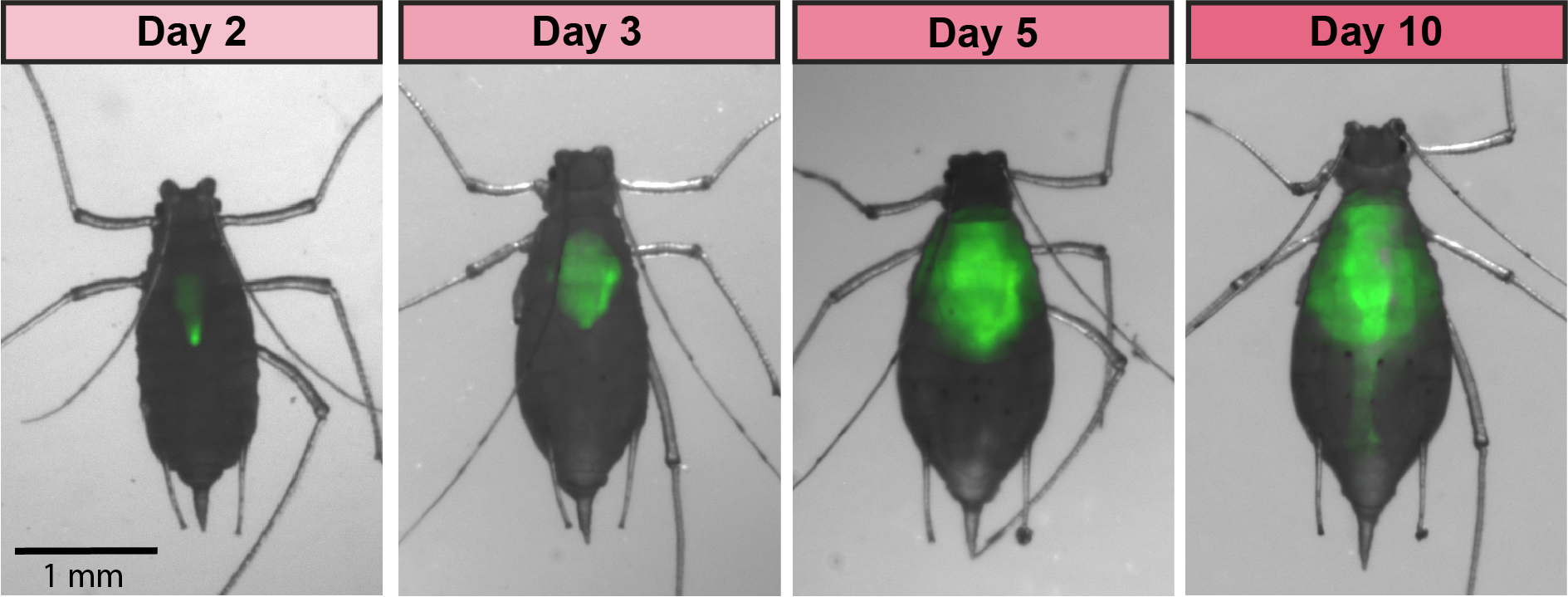
GFP-tagged symbiont colonizes the aphid gut
Aphids are plant pests and model systems for studying bacteria-insect symbioses. We have developed tools for engineering strains of
Serratia symbiotica that colonize the aphid gut. We are now using these tools to unravel how these strains, which can be pathogenic to their hosts, are related to strains of
S. symbiotica that live within insect cells, are inherited across aphid generations, and can benefit their aphid hosts. We have showed that cultured
S. symbiotica strains are capable of maternal transmission within aphids, suggesting that they possess a latent capacity to evolve a long-term symbiotic relationship with their hosts. We are also exploring applications of these engineered bacteria related to pest control.
 Researchers: Kate Elston
Researchers: Kate Elston,
Julie Perreau
 News: Turning Plant Pests into Helpers
News: Turning Plant Pests into Helpers
 Representative Publications
Representative Publications
- Elston et al. (2020) Applied and Environmental Microbiology bioRxiv
- Perreau et al. (2020) Submitted bioRxiv
Leafhoppers: Studying and Engineering Natural Nanomaterials
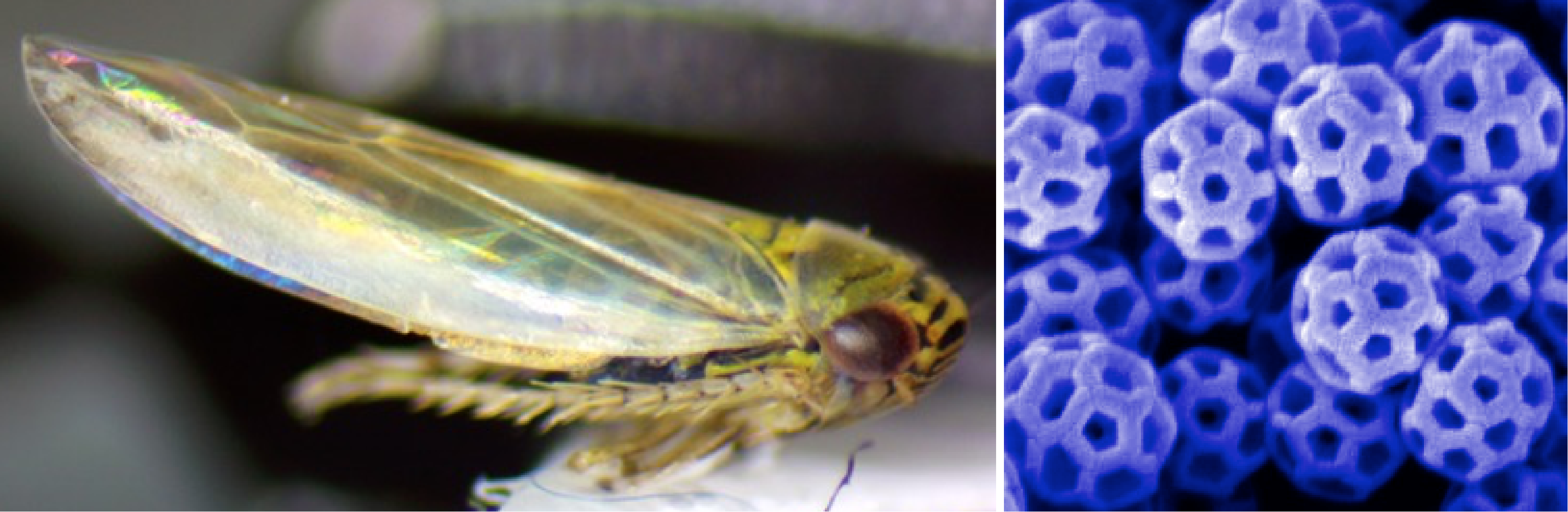
Leafhopper and SEM image of brochosomes on its wing
Leafhoppers produce unique nanostructures, called brochosomes, that they anoint onto their wings and sometimes eggs. Brochosomes have interesting materials properties, including superhydrophobicity and omnidirectional antireflectivity. We are characterizing natural variation in brochosome structures and performing comparative genomics to understand the molecular components that make up these structures, how they evolved, and their biological functions. We are also working towards genetically engineering symbionts of leafhoppers to create new tools for studying brochosomes and potentially enhancing their properties for various applications. The Barrick lab leads an interdisciplinary grant that includes the
Moran lab,
Jewett lab,
Schroeder lab,
Alleyne lab,
Milliron lab, and
Freeman lab to study brochosomes.
 Researchers: Sarah Bialik
Researchers: Sarah Bialik,
Kate Elston,
Peng Geng,
Elizabeth Robinson
 News: Tiny Insects Provide Inspiration for New Biomaterials
News: Tiny Insects Provide Inspiration for New Biomaterials
Preventing Evolutionary Failure in Synthetic Biology
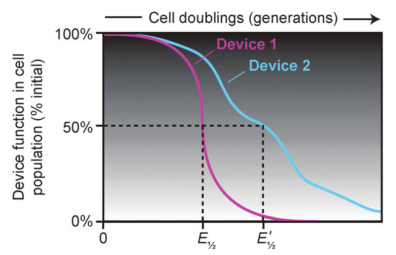
Evolutionary half-lives of biological devices
Synthetic biology applies engineering principles to create living systems with predictable and useful behaviors from collections of standardized genetic parts. However, living systems – unlike mechanical devices – inevitably evolve when their DNA sequences accumulate copying errors, often resulting in "broken" cells that no longer function as they were programmed. We are addressing this challenge by better characterizing how engineered cells evolve and using this information to design DNA sequences and host cells that are more robust against unwanted evolution. This work includes: (1) the development of the Evolutionary Failure Mode (EFM) Calculator software for identifying mutational hotspots in a designed DNA sequence; (2) using experimental evolution to identify "antimutator" variants of host organisms that lead to lower-than-natural mutation rates; and (3) designing genetic circuits that kill those cells within a population that are most likely to accumulate mutations.
 Researchers: Matt McGuffie
Researchers: Matt McGuffie,
Daniel Deatherage,
UT Austin iGEM Team
 Resources
Resources
 Representative Publications
Representative Publications
 Funding:
Funding: DARPA BRICS
Dynamics of Microbial Genome Evolution
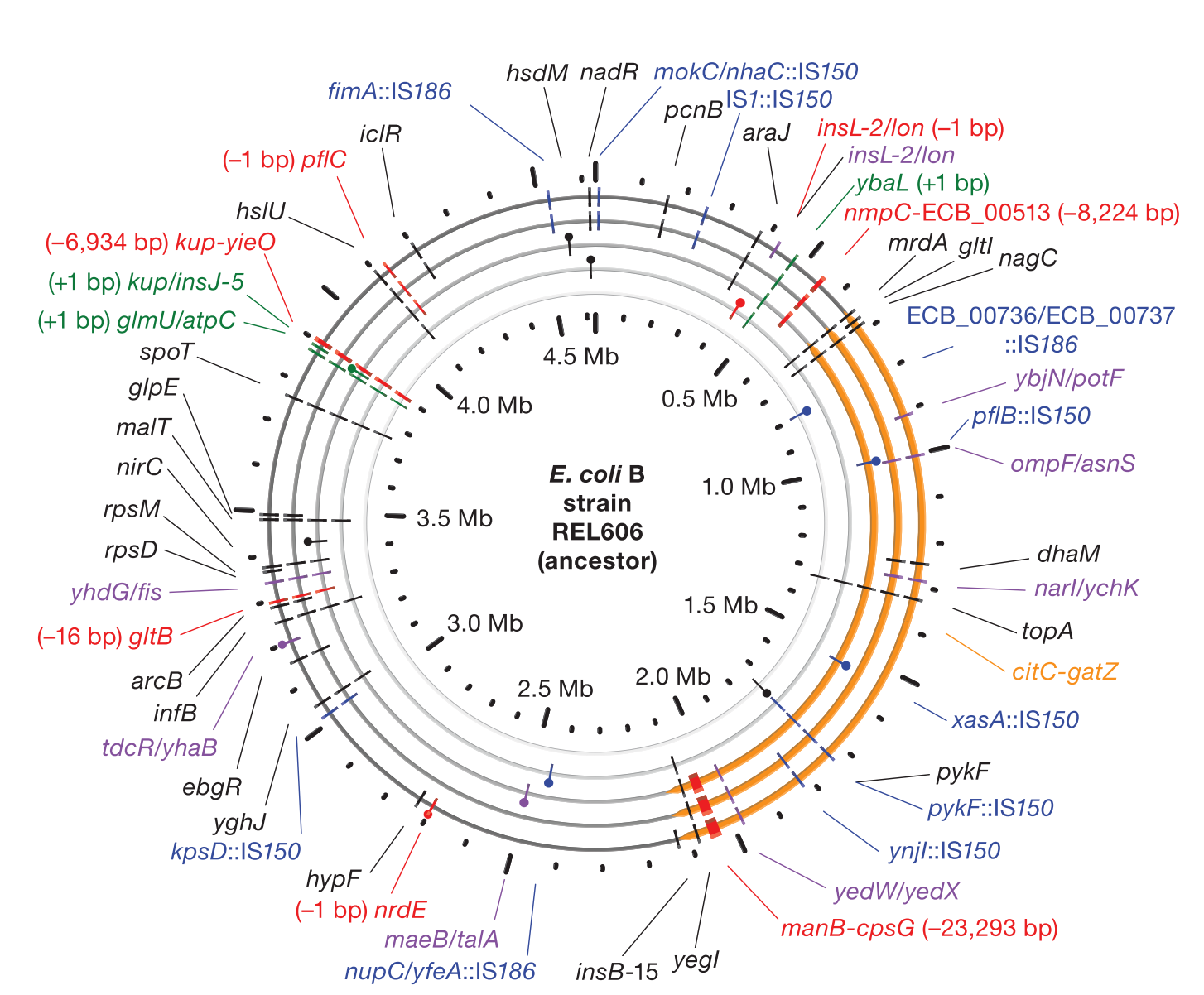
Accumulation of mutations in population Ara-1 of the LTEE over 20,000 generations of evolution
We develop the
breseq computational pipeline for identifying mutations in laboratory-evolved microbial genomes from next-generation sequencing data. We have used this tool to extensively study rates of genome evolution in the 30-year Lenski
long-term evolution experiment (LTEE) with
E. coli. We continue to develop
breseq so that it can be used for more additional applications related to strain engineering and medicine. For example, we are interested in how tracking rare variants within populations of microorganisms can anticipate further evolutionary trajectories and how this information might be used to better diagnose and treat disease.
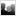 Researchers: Daniel Deatherage
Researchers: Daniel Deatherage
 Resources
Resources
 Representative Publications
Representative Publications
 Funding:
Funding: NIH K99/R00, NSF, NSF BEACON Center, CPRIT
Evolution and Engineering of Naturally Transformable Bacteria
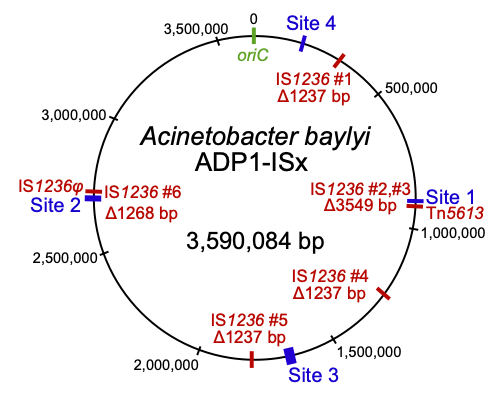
Insertion sequences (red) deleted in the transposon-free strain Acinetobacter baylyi ADP1-ISx
Naturally competent bacteria have expanded evolutionary potential because they can readily acquire new DNA from their environment. We are using experimental evolution of the model organism
Acinetobacter baylyi ADP1 to understand how horizontally acquired genes and mobile genetic elements become domesticated after their incorporation into a new genome and the broader effects of gene acquisition on the rest of the genome. We are also studying the role of chemical specificity in determining the fate of acquired DNA as either nutrition or genetic information. These bacteria also provide an improved platform for studying microbial genome engineering due to the ease of reconstructing mutations and introducing new genes. We are investigating sources of genetic instability in ADP1 and engineering a clean genome version of this strain by deleting transposable elements and prophages. This will promote the use of ADP1 in synthetic biology by reducing rates of mutations that lead to inactivation of introduced genes. Further, we are using ADP1 as a platform to understand the limits to streamlining bacterial genomes and identifying adaptations to overcome the fitness costs of reduced genomes.
 Researchers: Isaac Gifford
Researchers: Isaac Gifford
 Representative Publications
Representative Publications
 Funding:
Funding: Welch Foundation
Evolution and Engineering of Bacteriophages
Simulating engineered phage
The rise of multiple drug resistant pathogenic bacteria has raised the question of available, robust alternatives to antibiotics for disease treatment. Our lab is exploring new techniques for modifying bacteriophages for use in phage therapy. Evolution, engineering, and expansion of bacteriophage genomes has the potential to diversify target diseases for treatment, perform diagnostics for proactive disease prevention, and improve the efficacy of existing applications. We are working to accomplish these advancements through computational simulations and non-standard amino acid integration. The introduction of non-standard amino acids into the repertoire of protein production also brings insights into evolutionary outcomes that may not be possible in existing natural environments. Learning about evolution enabled by non-standard amino acids can lead to applications beyond the scope of viruses. We collaborate with the
Wilke lab to use their
Pinetree software for simulating phage gene expression and evolution.
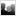 Researchers: Peng Geng
Researchers: Peng Geng,
Cameron Roots,
UT Austin iGEM Team
 Representative Publications
Representative Publications
 Funding:
Funding: NIH R01








Barrick Lab > ResearchInterests
 JeffreyBarrick, KateElston, SarahBialik, IsaacGifford, AlexaMorton, JuliePerreau, CameronRoots, GabrielSuarez
JeffreyBarrick, KateElston, SarahBialik, IsaacGifford, AlexaMorton, JuliePerreau, CameronRoots, GabrielSuarez
 Mol Biosciences
Mol Biosciences The LTEE
The LTEE iGEM team
iGEM team NGS course
NGS course