Barrick Lab :: Publications
Legend: Barrick Lab researchers *Equal contributions ^Corresponding author(s) Undergraduate researchers
| Preprints | |||
|---|---|---|---|
| 133. | *Morton, A.K., *Chaudhari, K., Matibag, B.D., Iyengar, V.B., Dullen, K.E., VanDieren, A.J., Parker, J.K., Mishler, D.M., ^Barrick, J.E. (2025) Refactored genetic parts for modular assembly of the E. coli MccV type I secretion system used to screen class II microcin candidates from plant-associated bacteria. bioRxiv DOI: 10.64898/2026.01.19.700402 | ||
| 132. | ^Navarro-Escalante, L., VanDieren, A.J., ^Barrick, J.E. (2025) Genetic biosensor for optimizing double-stranded RNA production by bacterial symbionts. bioRxiv DOI: 10.64898/2026.01.18.700149 | ||
| 131. | Ashraf, A H M Z., Weaver, K.K., Lariviere, P.J., ^Barrick, J.E. (2025) Natural competence promotes high rates of DNA transfer between strains of the core bee gut bacterium Snodgrassella alvi. bioRxiv DOI: 10.64898/2025.12.12.693966 | ||
| 130. | Gomez, J.B., Barrick, J.E., ^Waters, C.M. (2025) Phages use contingency loci as a bet-hedging strategy. bioRxiv DOI: 10.1101/2025.10.21.683753 | ||
| 129. | ^Lariviere, P.J., Ashraf, A H M Z., Gifford, I., Tanguma, S.L., Barrick, J.E., ^Moran, N.A. (2024) Virulence-linked adhesin drives mutualist colonization of the bee gut via biofilm formation. bioRxiv DOI: 10.1101/2024.10.14.618124 | ||
| 2025 | |||
| 128. | VanDieren, A.J., ^Barrick, J.E. (2025) Evolution in response to prophage activation attenuates the virulence of culturable Serratia symbiotica relatives of aphid endosymbionts. mBio 16:e0204125. DOI: 10.1128/mbio.02041-25 PMID: 40919815 | ||
| 127. | Fernandes, S.E., Ortega, H., Vaillancourt, M., Galdino, A.C.M., Stotland, A., Mun, K.S., Aguilar, D., Doi, Y., Lee, J.S., Burgener, E.B., Barrick, J.E., Schertzer J.W., ^Jorth, P. (2025). Evolutionary loss of an antibiotic efflux pump increases Pseudomonas aeruginosa quorum sensing mediated virulence in vivo. Nat. Commun. 16:8397. DOI: 10.1038/s41467-025-63284-7 PMID: 40998772 | ||
| 126. | ^Gifford, I., Vergis, M.R., ^Barrick, J.E. (2025) Genome evolution of Acinetobacter baylyi ADP1 during laboratory domestication: acquired mutations impact competence and metabolism. Appl. Env. Microbiol. 91:e0093625. DOI: 10.1128/aem.00936 PMID: 40827875 | ||
| 125. | Navarro-Escalante, L., Ashraf, A H M Z., Leonard, S.P., ^Barrick, J.E. (2025) Protecting honey bees through microbiome engineering. Curr. Opin. Insect Sci. 72:101416. DOI: 10.1016/j.cois.2025.101416 PMID: 40769317 | ||
| 124. | VanDieren, A.J., ^Barrick, J.E. (2025) UltraCAST: A Flexible All-In-One Suicide Vector for Modifying Bacterial Genomes Using a CRISPR-Associated Transposon. MicroPubl. Biol. DOI: 10.17912/micropub.biology.001721 | ||
| 123. | Zibbu, I., Wilke, C.O., ^Barrick, J.E. (2025). seabreeze: a pipeline for analyzing structural variation between bacterial genome assemblies. J. Open Source Softw. 10:8065. DOI: 10.21105/joss.08065 | ||
| 122. | Elias, E., Gifford, I., Didi, L., Fargeon, O., Arad, D., Cohen-Pavon, R., Sorek, G., Levin, L., Melamed, D., Aspit, L., Barrick, J.E., ^Bar-Yaacov, D. 2025. A-to-I mRNA editing recodes hundreds of genes in dozens of species and produces endogenous protein isoforms in bacteria. Nucleic Acids Res. 53:gkaf656. DOI: 10.1093/nar/gkaf656 PMID: 40650968 | ||
| 121. | Flatau, R., Krawczyk, A.I., Segoli, M., Barrick, J.E., ^Hawlena, H. (2025) Continuously high Wolbachia incidence in flea populations may result from dual-strain infections with divergent effects. Sci. Rep. 15:21720. DOI: 10.1038/s41598-025-09403-2 PMID: 40593332 | ||
| 120. | Chuong, J., Brown, K.W., Gifford, I., Mishler, D.M., ^Barrick, J.E. (2025) Engineered Acinetobacter baylyi ADP1-ISx cells are sensitive DNA biosensors for antibiotic resistance genes and a fungal pathogen of bats. ACS Synth. Biol. 14: 2488-2493. DOI: 10.1021/acssynbio.5c00360 PMID: 40579381 bioRxiv preprint | ||
| 119. | Lozzi, B., Adepoju, L., Espinoza, J.L., Padgen, M., Parra, M., Ricco, A., Castro-Wallace, S., Barrick, J.E., ^O’Rourke A. (2025) Simulated microgravity triggers a membrane adaptation to stress in E. coli REL606. BMC Microbiol. 25:362. DOI: 10.1186/s12866-025-04064-7 PMID: 40484963 | ||
| 118. | *Roots, C.T., *Hill, A.M., Wilke, C.O., ^Barrick, J.E. (2025) Modeling and measuring how codon usage modulates the relationship between burden and yield during protein overexpression in bacteria. J. Biol. Eng. 19:50. DOI: 10.1186/s13036-025-00521-z PMID: 40405198 | ||
| 117. | Lay, C.G., Burks, G.R., Li, Z., Barrick, J.E., Schroeder, C.M., Karim, A.S., ^Jewett, M.C. (2025) Cell-free expression of soluble leafhopper proteins from brochosomes. ACS Synth. Biol. 21: 987-994. DOI: 10.1021/acssynbio.4c00773 PMID: 40052868 | ||
| 2024 | |||
| 116. | Roots, C.T., ^Barrick, J.E. (2024) CryptKeeper: a negative design tool for reducing unintentional gene expression in bacteria. Synth. Biol. 9:ysae018. DOI: 10.1093/synbio/ysae018 PMID: 39722801 | ||
| 115. | Rodríguez‑Pastor, R. Knossow N., Shahar, N, Hasik, A.Z., Deatherage, D.E., Gutiérrez, R., Harrus, S.,Zaman, L., Lenski, R.E., ^Barrick, J.E., ^Hawlena, H. (2024) Pathogen contingency loci and the evolution of host specificity: Simple sequence repeats mediate Bartonella adaptation to a wild rodent host. PLoS Pathogens 20:e1012591. DOI: 10.1371/journal.ppat.1012591 PMID: 39348417 | ||
| 114. | *Gifford, I., *Suárez G.A., ^Barrick, J.E. (2024) Evolution recovers the fitness of Acinetobacter baylyi strains with large deletions through mutations in deletion-specific targets and global post-transcriptional regulators. PLoS Genetics 20:e1011306. DOI: 10.1371/journal.pgen.1011306 PMID: 39283914 | ||
| 113. | ^Lariviere, P.J., Ashraf, A H M Z., Navarro-Escalante, L., Leonard, S.P., Miller, L.G., Moran, N.A. ^Barrick, J.E. (2024) One-step genome engineering in bee gut bacterial symbionts. mBio 15:e0139224. DOI: 10.1128/mbio.01392-24 PMID: 39105596 | ||
| 112. | *Radde, N., *Mortensen, G.A., Bhat, D., Shah, S., Clements, J.J., Leonard, S.P., McGuffie, M.J., Mishler, D.M., ^Barrick, J.E. (2024) Measuring the burden of hundreds of BioBricks defines an evolutionary limit on constructability in synthetic biology. Nat. Commun. 15: 6242. DOI: 10.1038/s41467-024-50639-9 PMID: 39048554 | ||
| 111. | McGuffie, M.J., ^Barrick, J.E. (2024) Identifying widespread and recurrent variants of genetic parts to improve annotation of engineered DNA sequences. PLoS ONE 19:e0304164. DOI: 10.1371/journal.pone.0304164 PMID: 38805426 | ||
| 110. | uz-Zaman, Md. H., D'Alton, S., Barrick, J.E., ^Ochman H. (2024) Promoter recruitment drives the emergence of proto-genes in a long-term evolution experiment with Escherichia coli. PLoS Biol. 22:e3002418. DOI: 10.1371/journal.pbio.3002418 PMID: 38713714 | ||
| 109. | Diaz Lluberes, I.A., Ahmido, T., Fernando, P.U.A.I., Kosgei, G.K., Bayliff, K.W., Dwenger IV, J.H., Dolocan, A., Barrick, J.E., Smith, C.B. (2024) Exploring the extended spectral signature and remote sensing capabilities of lanthanide enriched arrays of brochosomes. In: W.M. Shensky III, I. Rau, O. Sugihara (eds.). Organic Photonic Materials and Devices XXVI 12883:1288304. DOI: 10.1117/12.3001089 | ||
| 2023 | |||
| 108. | Thuronyi, B W., DeBenedictis, E.A., ^Barrick, J.E. (2023) No assembly required: Time for stronger, simpler publishing standards for DNA sequences. PLoS Bio. 21:e3002376. DOI: 10.1371/journal.pbio.3002376 PMID: 37971964  |
||
| 107. | Rodríguez-Pastor, R., Hasik, A.Z., Knossow, N., Bar-Shira, E., Shahar, N., Gutiérrez, R., Zaman, L., Harrus, S., Lenski, R.E., Barrick, J.E., ^Hawlena, H. (2023) Bartonella infections are prevalent in rodents despite efficient immune responses. Parasites Vectors 16:315. DOI: 10.1186/s13071-023-05918-7 PMID: 37667323 | ||
| 106. | ^Barrick, J.E., ^Blount, Z.D., Lake, D.M., Dwenger IV, J.H., Chavarria-Palma, J.E., Izutsu, M., Wiser, M.J. (2023) Daily transfers, archiving populations, and measuring fitness in the long-term evolution experiment with Escherichia coli. J. Vis. Exp. 198:e65342. DOI: 10.3791/65342 PMID: 37607082 | ||
| 105. | *Elston, K.M., *Phillips, L.E., Leonard, S.P., Young, E., Holley, J.C., Ahsanullah, T., McReynolds, B., Moran, N.A., ^Barrick, J.E. (2023) The Pathfinder plasmid toolkit for genetically engineering newly isolated bacteria enables the study of Drosophila-colonizing Orbaceae. ISME Commun. 3:9. DOI: 10.1038/s43705-023-00255-3 PMID: 37225918 | ||
| 104. | ^Banerjee, P., Burks, G.R., Bialik, S.B., Bello, E., Alleyne, M., Freeman, B.D., Barrick, J.E., Schroeder C.M., Milliron, D.J. (2023) Nanostructure-derived anti-reflectivity in leafhopper brochosomes. Adv. Photonics Res. 202200343. DOI: 10.1002/adpr.202200343 ChemRxiv | ||
| 103. | Elston, K.M., Maeda, G.P., Perreau, J., ^Barrick, J.E. (2023) Addressing the challenges of symbiont-mediated RNAi in aphids. PeerJ 11:e14961. DOI: 10.7717/peerj.14961 PMID: 36874963 | ||
| 102. | Burks, G.R., Yao, L., Kalutantirige, F.C., Gray, K.J., Bello, E., Rajagopalan, S., Bialik, S.B., Barrick, J.E., Alleyne, M., Chen, Q., ^Schroeder C.M. (2023) Electron tomography and machine learning for understanding the highly ordered structure of leafhopper brochosomes. Biomacromolecules 24:190-200. DOI: 10.1021/acs.biomac.2c01035 PMID: 36516996 | ||
| 101. | *Lariviere, P.J., *Leonard, S.P., Horak, R.D., Powell, J.E., ^Barrick, J.E. (2023) Honey bee functional genomics using symbiont-mediated RNAi. Nat. Protoc. 18:902-928. DOI: 10.1038/s41596-022-00778-4 PMID: 36460809 Preprint | ||
| 2022 | |||
| 100. | Maritan, E., Gallo, M., Srutkova, D., Jelinkova, A., Benada, O., Kofronova, O., Silva-Soares, N.F., Hudcovic, T., Gifford, I., Barrick, J.E., Schwarzer, M, Martino, M. E. (2022) Gut microbe Lactiplantibacillus plantarum undergoes different evolutionary trajectories between insects and mammals. BMC Biol. 20:290. DOI: 10.1186/s12915-022-01477-y PMID: 36575413 | ||
| 99. | Li, Z., Li, Y, Xue A.Z., Dang V., Holmes, V.R., Johnston, J.S., Barrick, J.E., ^Moran, N.A. (2022) The genomic basis of evolutionary novelties in a leafhopper. Mol. Biol. Evol. 39:msac184. DOI: 10.1093/molbev/msac184 PMID: 36026509 | ||
| 98. | Rodríguez-Pastor, R., Shafran, Y., Knossow, N., Gutiérrez, R. Harrus, S., Zaman, L., Lenski, R. E., Barrick, J. E., ^Hawlena, H. (2022) A road map for in vivo evolution experiments with blood-borne parasitic microbes. Mol. Ecol. Resour. 22:2843-2859. DOI: 10.1111/1755-0998.13649 PMID: 35599628 | ||
| 97. | Gómez-Garzón, C, Barrick, J.E., ^Payne, S.M. (2022) Disentangling the evolutionary history of Feo, the major ferrous iron transport system in bacteria. mBio 13:e03512-21. DOI: 10.1128/mbio.03512-21 PMID: 35012344 | ||
| 96. | Elston, K.M., Leonard, S.P., Geng, P., Bialik, S.B., Robinson, E., ^Barrick, J.E. (2022) Engineering insects from the endosymbiont out. Trends Microbiol. 30:79-96. DOI: 10.1016/j.tim.2021.05.004 PMID: 34103228  |
||
| 2021 | |||
| 95. | Deatherage, D.E., ^Barrick, J.E. (2021) High-throughput characterization of mutations in genes that drive clonal evolution using multiplex adaptome capture sequencing. Cell Syst. 12:79-96. DOI: 10.1016/j.cels.2021.08.011 PMID: 34536379 bioRxiv  |
||
| 94. | Roots, C.T., Lukasiewicz, A., ^Barrick, J.E. (2021) OSTIR: open source translation initiation rate prediction. J. Open Source Softw. 6:3362. DOI: 10.21105/joss.03362 | ||
| 93. | Borin, J.M., Avrani, S., Barrick, J.E., Petrie, K.L., ^Meyer, J.R. (2021) Coevolutionary phage training leads to greater bacterial suppression and delays the evolution of phage resistance. Proc. Natl. Acad. Sci. U.S.A. 118:e2104592118. DOI: 10.1073/pnas.2104592118 PMID: 34083444 | ||
| 92. | McGuffie, M.C., ^Barrick, J.E. (2021) pLannotate: engineered plasmid annotation. Nucleic Acids Res. 49:W516-W522. DOI: 10.1093/nar/gkab374 PMID: 34019636
 |
||
| 91. | ^Perreau, J., Patel, D.J., Anderson, H., Maeda, G.P, Elston, K.M., Barrick, J.E., Moran, N.A. (2021) Vertical transmission at the pathogen-symbiont interface: Serratia symbiotica and aphids. mBio 12:e00359-21. DOI: 10.1128/mBio.00359-21 PMID: 33879583 | ||
| 90. | Consuegra, J., Gaffé, J., Lenski, R. E., Hindré, T., Barrick, J. E., Tenaillon, O., Schneider, D. (2021) Insertion-sequence-mediated mutations both promote and constrain evolvability during a long-term experiment with bacteria. Nat. Commun. 12:980. DOI: 10.1038/s41467-021-21210-7 PMID: 33579917
|
||
| 89. | *Bazurto, J.V., *Riazi, S., D’Alton, S., Deatherage, D.E., Bruger, E.L., Barrick, J.E., ^Marx, C.J. (2021) Global transcriptional response of Methylorubrum extorquens to formaldehyde stress expands the role of EfgA and is distinct from antibiotic translational inhibition. Microorganisms 9:347. DOI: 10.3390/microorganisms9020347 PMID: 33578755 | ||
| 88. | ^Card, K.J., Thomas, M.D., Graves, J.L., Barrick, J.E., Lenski, R.E. (2021) Genomic evolution of antibiotic resistance is contingent on genetic background following a long-term experiment with Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 118: e2016886118. DOI: 10.1073/pnas.2016886118 PMID: 33441451 | ||
| 87. | *Gifford, I., *Dasgupta, A., ^Barrick, J.E. (2021) Rates of gene conversions between Escherichia coli ribosomal operons. G3 11:jkaa002. DOI: 10.1093/g3journal/jkaa002 PMID: 33585862 | ||
| 86. | Elston, K.M., Perreau, J., Maeda, G.P., Moran, N.A., ^Barrick, J.E. (2021) Engineering a culturable Serratia symbiotica strain for aphid paratransgenesis. Appl. Env. Microbiol. 87:e02245-20. DOI: 10.1128/AEM.02245-20 PMID: 33277267 |
||
| 2020 | |||
| 85. | *Sher, A.A., *Jerome, J.P., Bell, J.A., Yu, J., Kim, H.Y., Barrick, J.E., ^Mansfield, L.S. (2020) Experimental evolution of Campylobacter jejuni leads to loss of motility, rpoN (σ54) deletion and genome reduction. Front. Microbiol. 11:579989 DOI: 10.3389/fmicb.2020.579989 PMID: 33240235 | ||
| 84. | ^Barrick, J.E. (2020) Limits to predicting evolution: Insights from a long-term experiment with Escherichia coli. In: W. Banzhaf, B.H.C. Cheng, K. Deb, K.E. Holekamp, R.E. Lenski, C. Ofria, R.T. Pennock, W.F. Punch, D.J. Whittaker (eds.). Evolution in Action: Past, Present and Future: A Festschrift in Honor of Erik D. Goodman. pp 63-76. Springer Series on Genetic and Evolutionary Computation. Cham, Switzerland: Springer. Full Text DOI: 10.1007/978-3-030-39831-6_7 | ||
| 83. | ^Barrick, J.E., Deatherage D.E., ^Lenski, R.E. (2020) A test of the repeatability of measurements of relative fitness in the long-term evolution experiment with Escherichia coli. In: W. Banzhaf, B.H.C. Cheng, K. Deb, K.E. Holekamp, R.E. Lenski, C. Ofria, R.T. Pennock, W.F. Punch, D.J. Whittaker (eds.). Evolution in Action: Past, Present and Future: A Festschrift in Honor of Erik D. Goodman. pp 77-89. Springer Series on Genetic and Evolutionary Computation. Cham, Switzerland: Springer. Full Text DOI: 10.1007/978-3-030-39831-6_8 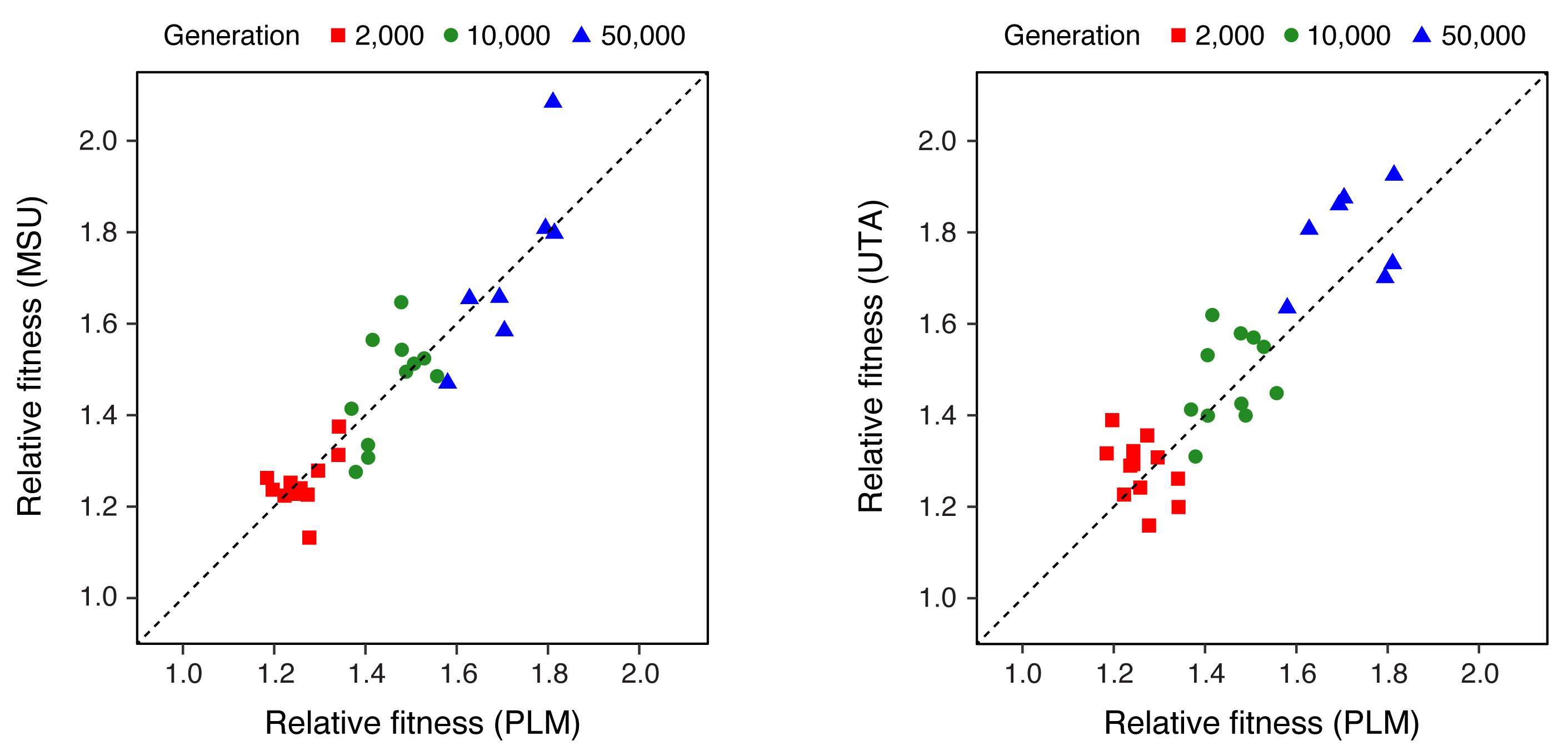 |
||
| 82. | ^*Blount, Z.D., ^*Maddamsetti, R., *Grant, N.A., Ahmed, S.T., Jagdish, T., Sommerfeld, B.A., Tillman, A., Moore, J., Slonczewski, J.L., Barrick, J.E., Lenski, R.E. (2020) Genomic and phenotypic evolution of Escherichia coli in a novel citrate-only resource environment. eLife 9:e55414. Full Text PMID: 32469311 | ||
| 81. | Suárez, G.A., Dugan, K.R., Renda, B.A., Leonard, S.P., Gangavarapu, L.S., ^Barrick, J.E. (2020) Rapid and assured genetic engineering methods applied to Acinetobacter baylyi ADP1 genome streamlining. Nucleic Acids Res. 48:4585–4600. Full Text PMID: 32232367 | ||
| 80. | Leonard, S.P., Powell, J.E., Perutka, J., Geng, P., Heckmann, L.C., Horak, R.D., Davies, B.W., Ellington, A.D., ^Barrick, J.E., and ^Moran, N.A. (2020) Engineered symbionts activate honey bee immunity and limit pathogens. Science 367:573–576. DOI: 10.1126/science.aax9039 PMID: 32001655  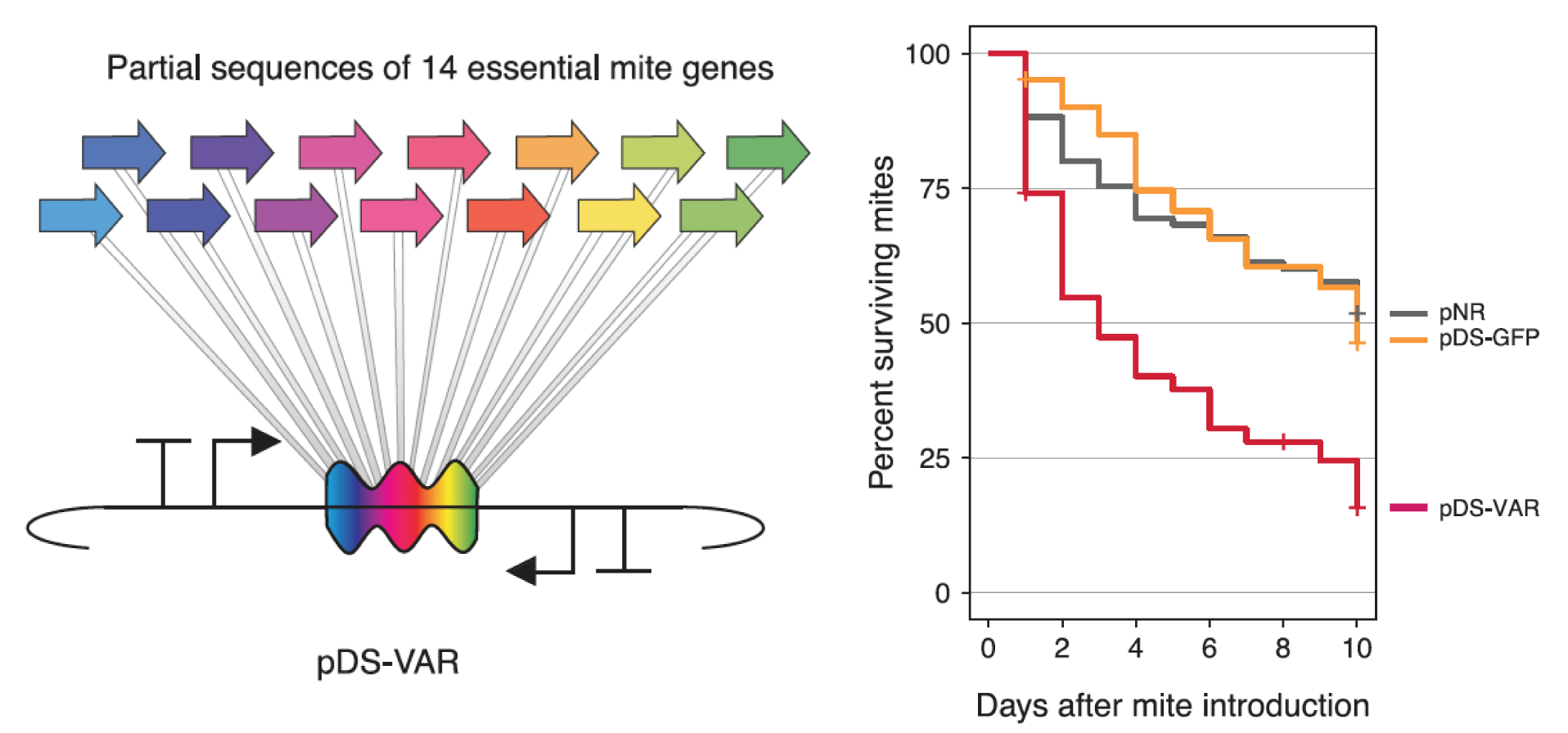 |
||
| 2019 | |||
| 79. | Zhang, X., Deatherage, D.E., Zheng, H., Georgoulis, S.J., ^Barrick, J.E. (2019) Evolution of satellite plasmids can prolong the maintenance of newly acquired accessory genes in bacteria. Nat. Comm. 10:5809. DOI: 10.1038/s41467-019-13709-x Full Text, PMID: 31863068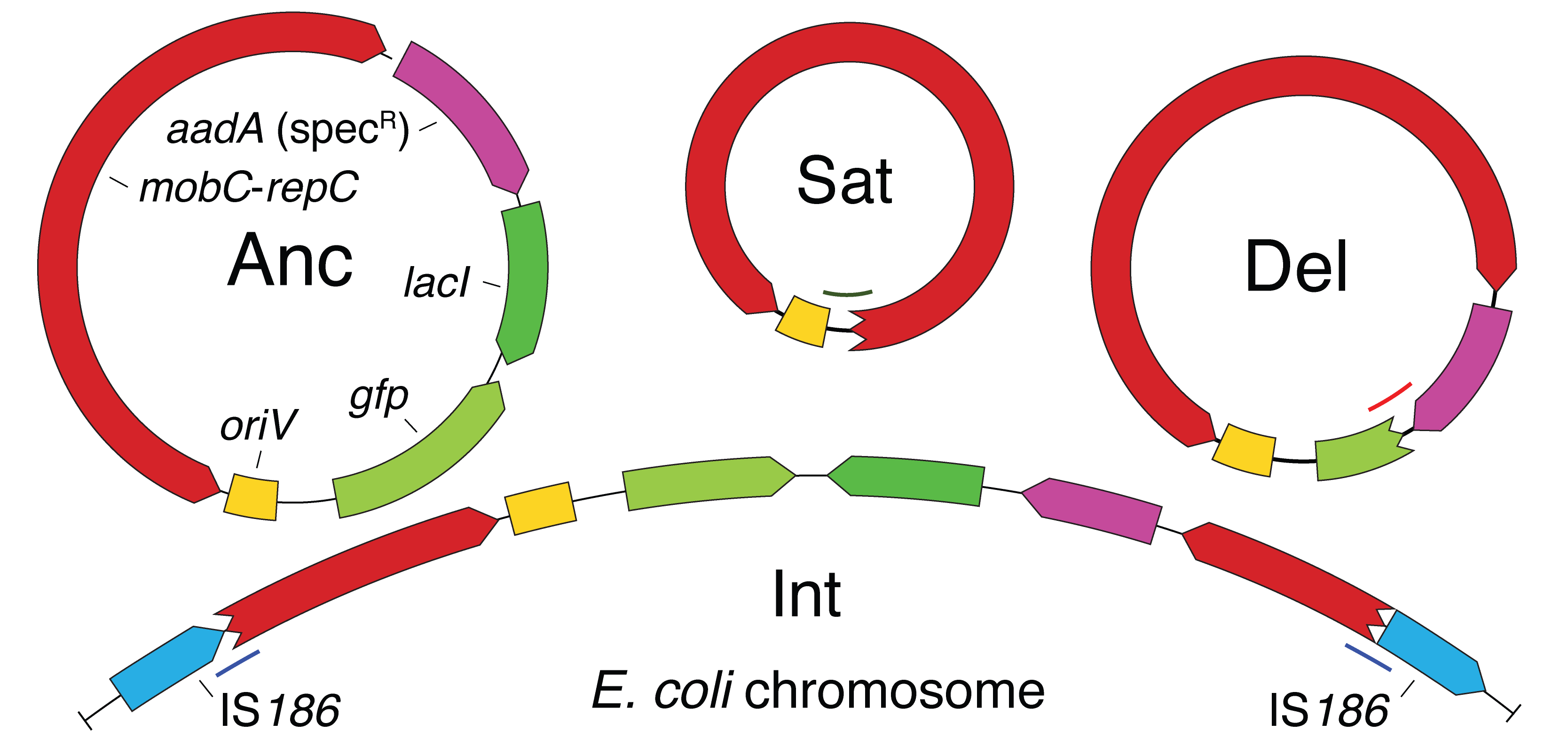 |
||
| 78. | Gutierrez, A.E., Shah, P., Rex, A.E., Nguyen, T.C., Kenkare, S.P., ^Barrick, J.E., ^Mishler, D.M. (2019) Bioassay for determining the concentrations of caffeine and individual methylxanthines in complex samples. Appl. Env. Microbiol. 85:e01965-19. PMID: 31540989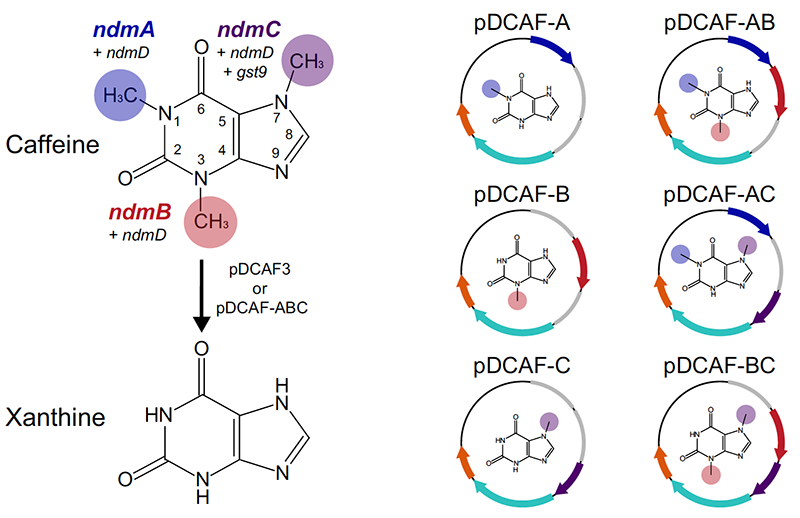 |
||
| 77. | Geng, P., Leonard, S.P., Mishler, D.M., ^Barrick, J.E. (2019) Synthetic genome defenses against selfish DNA elements stabilize engineered bacteria against evolutionary failure. ACS Synth. Biol. 8:521-531. DOI: 10.1021/acssynbio.8b00426, Preprint, PMID: 30703321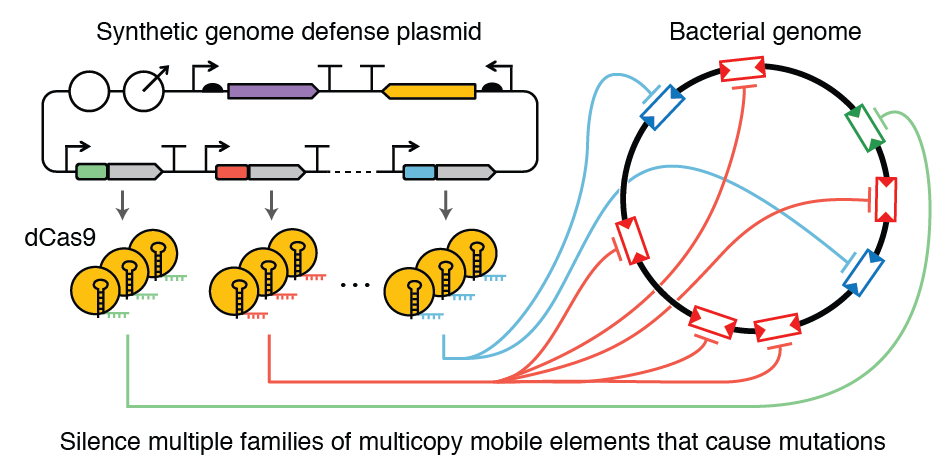 |
||
| 2018 | |||
| 76. | Deatherage, D.E., Leon, D., Rodriguez, Á.E., Omar, S., ^Barrick, J.E. (2018) Directed evolution of Escherichia coli with lower-than-natural plasmid mutation rates. Nucleic Acids Res. 46:9236-9250. DOI: 10.1093/nar/gky751, PMID: 30137492  |
||
| 75. | *McGuffey, J.C., *Leon, D., Dhanji, E.Z., Mishler, D.M., ^Barrick, J.E. (2018) Bacterial production of gellan gum as a do-it-yourself alternative to agar. J. Microbiol. Biol. Educ. 19:182-184. Full Text, PMID: 29983852 |
||
| 74. | Leonard, S.P., Perutka, J., Powell, J.E., Geng, P., Richhart, D., Byrom, M., Kar, S., Davies, B.W., Ellington, A.D., ^Moran, N.A., ^Barrick, J.E. (2018) Genetic engineering of bee gut microbiome bacteria with a toolkit for modular assembly of broad-host-range plasmids. ACS Synth. Biol. 7:1279-1290. DOI: 10.1021/acssynbio.7b00399 PMID: 29608282 |
||
| 73. | Leon, D., D’Alton, S., Quandt, E.M., ^Barrick, J.E. (2018) Innovation in an E. coli evolution experiment is contingent on maintaining adaptive potential until competition subsides. PLoS Genet. 14:e1007348. DOI: 10.1371/journal.pgen.1007348 PMID: 29649242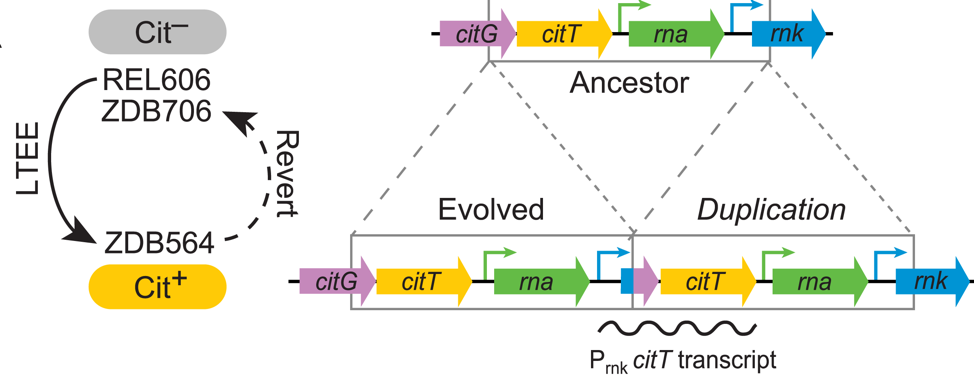 |
||
| 2017 | |||
| 72. | Good, B.H.*, McDonald, M.J.*, Barrick, J.E., Lenski R.E., Desai, M.M. (2017) The dynamics of molecular evolution over 60,000 generations. Nature 551:45-50. DOI: 10.1038/nature24287 PMID: 29045390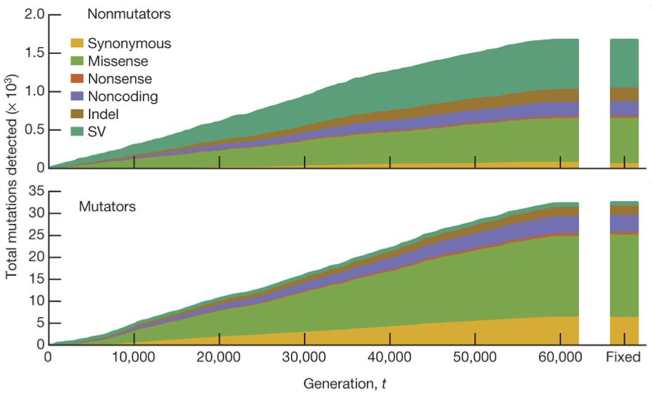 |
||
| 71. | Bull, J.J.*, Barrick, J.E.^* (2017) Arresting evolution. Trends Genet. 33:910-920. PMID: 29029851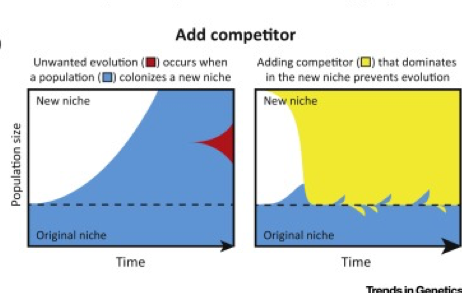 |
||
| 70. | ^Barrick, J.E. (2017) In future (cell) generations. “Voices” piece in What is the role of circuit design in the advancement of synthetic biology? Part 2. Cell Syst. 4:467-477. PMID: 28544877 | ||
| 69. | Suarez, G.A., Renda, B.A., Dasgupta, A., Barrick, J.E.^ (2017) Reduced mutation rate and increased transformability of transposon-free Acinetobacter baylyi ADP1-ISx. Appl. Env. Microbiol. 83: e01025-17. DOI: 10.1128/AEM.01025-17 PMID: 28667117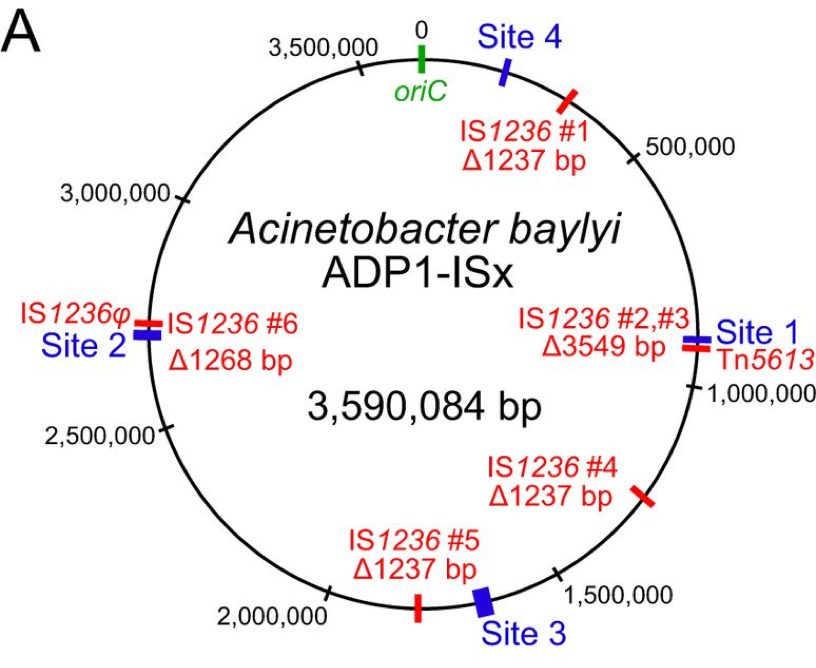 |
||
| 68. | Brown, C.W., Sridhara, V., Boutz, D.R., Person, M.D., Marcotte, E.M. Barrick, J.E., Wilke, C.O.^ (2017) Large-scale analysis of post-translational modifications in E. coli under glucose-limiting conditions. BMC Genomics 18:301. PMID: 28412930 | ||
| 67. | Caglar, M.U., Houser, J.R., Barnhart, C.S., Boutz, D.R., Carroll, S.M., Dasgupta, A., Lenoir, W.F., Smith, B.L., Sridhara, V., Sydykova, D.K., Vander Wood, D., Marx, C.J., Marcotte, E.M.^, Barrick, J.E.^, Wilke, C.O.^ (2017) The E. coli molecular phenotype under different growth conditions. Sci. Rep. 7:45303. PMID: 28417974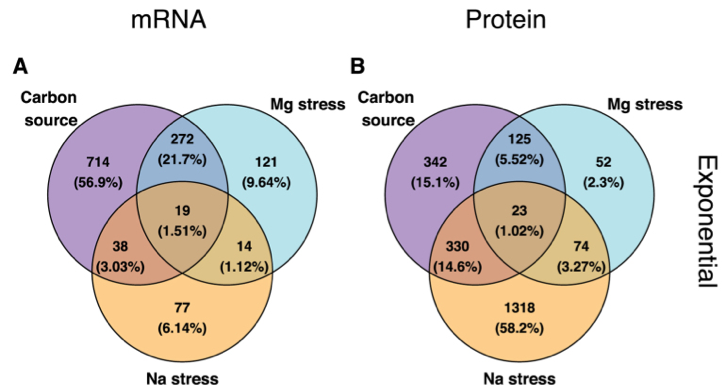 |
||
| 66. | Deatherage, D.E., Kepner, J.L., Bennett, A.F., Lenski, R.E.^, Barrick, J.E.^ (2017) Specificity of genome evolution in experimental populations of Escherichia coli evolved at different temperatures. Proc. Natl. Acad. Sci. U.S.A. 114:E1904-E1912. PMID: 28202733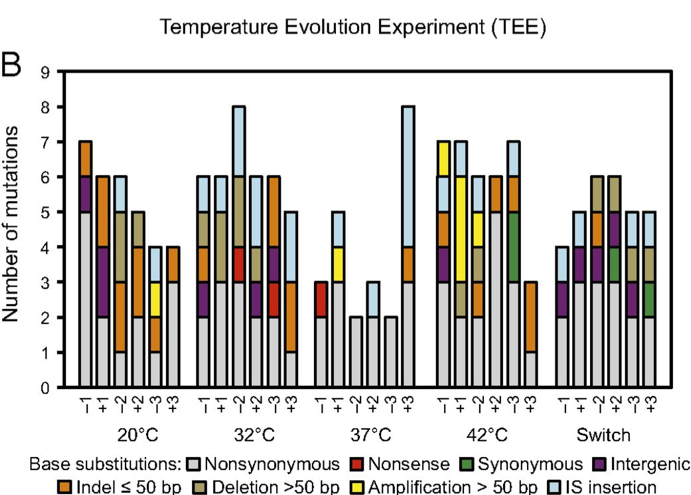 |
||
| 65. | Monk, J.W., Leonard, S.P., Brown, C.W., Hammerling, M.J., Mortensen, C., Gutierrez, A.E., Shin, N.Y., Watkins, E., Mishler, D.M.^, Barrick, J.E.^ (2017) Rapid and inexpensive evaluation of nonstandard amino acid incorporation in Escherichia coli. ACS Synth. Biol. 6:45-54. PMID: 27648665 |
||
| 2016 | |||
| 64. | Renda, B.A., Chan, C., Parent, K.N., Barrick, J.E.^ (2016) Emergence of a competence reducing filamentous phage from the genome of Acinetobacter baylyi ADP1. J. Bacteriol. 198:3209-3219. PMID: 27645387 | ||
| 63. | Tenaillon, O.*, Barrick, J.E.*, Ribeck, N., Deatherage, D.E., Blanchard, J.L, Dasgupta, A., Wu, G.C., Wielgoss, S., Cruveiller, S., Medigue, C., Schneider, D., Lenski, R.E.^ (2016) Tempo and mode of genome evolution in a 50,000-generation experiment. Nature 536:165-170. PMID: 27479321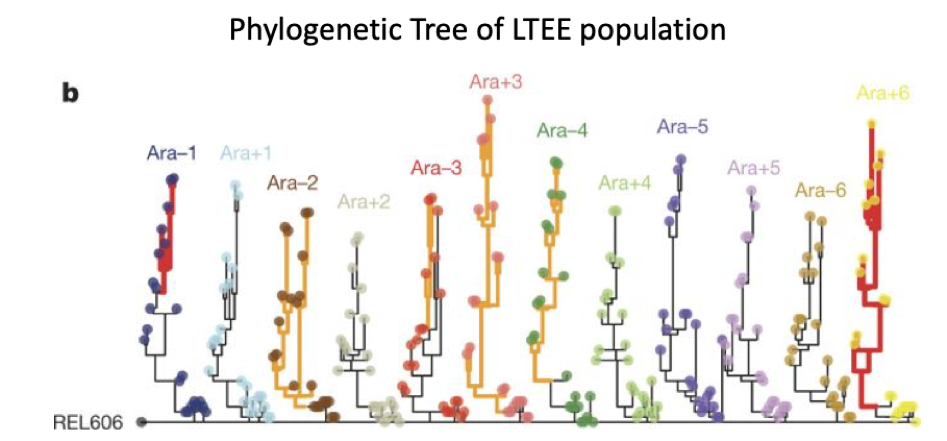 |
||
| 62. | Hammerling, M.J., Gollihar, J., Mortensen, C., Alnahhas, R.N., Ellington, A.D., Barrick, J.E. (2016) Expanded genetic codes create new mutational routes to rifampicin resistance in Escherichia coli. Mol. Biol. Evol. 33:2054-2063. PMID: 27189550 | ||
| 2015 | |||
| 61. | Quandt, E.M., Gollihar, J., Blount, Z.D., Ellington, A.D., Georgiou, G., Barrick, J.E. (2015) Fine-tuning citrate synthase flux potentiates and refines metabolic innovation in the Lenski evolution experiment. eLife 4:e09696. DOI: 10.7554/eLife.09696 PMID: 26465114 |
||
| 60. | Houser, J.R., Barnhart, C., Boutz, D.R., Carroll, S.M., Dasgupta, A., Michener, J.K., Needham, B.D., Papoulas, O., Sridhara, V., Sydykova, D.K., Marx, C.J., Trent, S.M., Barrick, J.E.^, Marcotte, E.M.^, Wilke, C.O.^ (2015) Controlled measurement and comparative analysis of cellular components in E. coli reveals broad regulatory changes in response to glucose starvation. PLoS Comput. Biol. 11:e1004400. PMID:26275208 | ||
| 59. | Maddamsetti, R., Hatcher, P.J., Cruveiller, S., Médigue, C., Barrick, J.E., Lenski, R.E. (2015) Synonymous genetic variation in natural isolates of Escherichia coli does not predict where synonymous substitutions occur in a long-term experiment. Mol. Biol. Evol. 32:2897-2904. PMID:26199375 | ||
| 58. | Jack, B.R., Leonard, S.P., Mishler, D.M., Renda, B.A., Leon, D., Suárez, G.A., Barrick, J.E. (2015) Predicting the genetic stability of engineered DNA sequences with the EFM Calculator. ACS Synth. Biol. 4:939-943. DOI: 10.1021/acssynbio.5b00068 PMID:26096262 | ||
| 57. | Perry, E.B.^, Barrick, J.E., Bohannan, B.J.M. (2015) The molecular and genetic basis of repeatable coevolution between Escherichia coli and bacteriophage T3 in a laboratory microcosm. PLoS ONE 10:e0130639. PMID:26114300. | ||
| 56. | Quandt, E.M., Summers, R.M., Subramanian, M.V., Barrick, J.E.^ (2015) Draft genome sequence of the bacterium Pseudomonas putida CBB5, which can utilize caffeine as a sole carbon and nitrogen source. Genome Announc. 3:e00640-15. PMID:26067973 | ||
| 55. | Maddamsetti, R., Lenski, R.E., Barrick, J.E. (2015) Adaptation, clonal interference, and frequency-dependent interactions in a long-term evolution experiment with Escherichia coli. Genetics 200:619-631. PMID:25911659 |
||
| 54. | Graves, J.L.^, Tajkarimi, M., Cunningham, Q., Campbell, A., Nonga, H., Harrison, S.H., Barrick, J.E. (2015) Rapid evolution of silver nanoparticle resistance in Escherichia coli. Front. Genet. 6:42. PMID:25741363 | ||
| 53. | Renda, B.A., Dasgupta, A., Leon, D., Barrick, J.E. (2015) Genome instability mediates the loss of key traits by Acinetobacter baylyi ADP1 during laboratory evolution. J. Bacteriol. 197:872-881. PMID:25512307 | ||
| 52. | Deatherage, D.E., Traverse, C.C., Wolf, L.N., Barrick, J.E. (2015) Detecting rare structural variation in evolving microbial populations from new sequence junctions using breseq. Front. Genet. 5:468. PMID:25653667 | ||
| 2014 | |||
| 51. | Sridhara, V., Meyer, A.G., Rai, P., Barrick, J.E., Ravikumar, P., Ségre, D., Wilke, C.O. (2014) Predicting growth conditions from internal metabolic fluxes in an in-silico model of E. coli. PLoS ONE 9:e114608. PMID:25502413 | ||
| 50. | Alnahhas, R.N.*, Slater, B.*, Huang, Y., Mortensen, C., Monk, J.W., Okasheh, Y., Howard, M.D., Gottel, N.R., Hammerling, M.J.^, Barrick, J.E.^ (2014) The case for decoupling assembly and submission standards to maintain a more flexible registry of biological parts. J. Biol. Eng. 8:28. PMID:25525459 | ||
| 49. | Barrick, J.E., Colburn, G., Deatherage D.E., Traverse, C.C., Strand, M.D., Borges, J.J., Knoester, D.B., Reba, A., Meyer, A.G. (2014) Identifying structural variation in haploid microbial genomes from short-read resequencing data using breseq. BMC Genomics 15:1039. PMID:25432719 | ||
| 48. | Raeside, C., Gaffé, J., Deatherage, D.E., Tenaillon, O., Briska, A.M., Ptashkin, R.N., Cruveiller, S., Médigue, C., Lenski, R.E., Barrick, J.E., Schneider, D. (2014) Large chromosomal rearrangements during a long-term evolution experiment with Escherichia coli. mBio 5:e01377-14. PubMed:25205090 | ||
| 47. | Deatherage, D.E., Barrick, J.E. (2014) Identification of mutations in laboratory-evolved microbes from next-generation sequencing data using breseq. Methods Mol. Biol. 1151:165-188. DOI: 10.1007/978-1-4939-0554-6_12 PMID: 24838886 | ||
| 46. | Renda, B.A.*, Hammerling, M.J.*, Barrick, J.E. (2014) Engineering reduced evolutionary potential for synthetic biology. Mol. Biosyst. 10:1668-1678. DOI: 10.1039/c3mb70606k PubMed:24556867 | ||
| 45. | Hammerling, M.J., Ellefson, J.W., Boutz, D.R., Marcotte, E.M., Ellington, A.D., Barrick, J.E. (2014) Bacteriophages use an expanded genetic code on evolutionary paths to higher fitness. Nat. Chem. Biol. 10:178-180. PubMed:24487692 | ||
| 44. | Quandt, E.M., Deatherage, D.E., Ellington, A.D., Georgiou, G., Barrick, J.E. (2014) Recursive genomewide recombination and sequencing reveals a key refinement step in the evolution of a metabolic innovation in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 111:2217-2222. PubMed:24379390 |
||
| 2013 | |||
| 43. | Hammerling, M.J., Gottel, N.R., Alnahhas, R.N., Slater, B., Huang, Y., Okasheh, Y., Howard, M., Mortensen, C., Monk, J., Detelich, M., Lannan, R.S., Pitaktong, A., Weaver, E., Das, S., Barrick, J.E. (2013) BBF RFC95: Open Sequence Initiative: a part submission standard to complement modern DNA assembly techniques. «BioBricks Foundation» «DSpace» | ||
| 42. | Barrick, J.E., Lenski, R.E. (2013) Genome dynamics during experimental evolution. Nat. Rev. Genet. 14:827-834. «PubMed» | ||
| 41. | Summers, R.M., Seffernick, J.L., Quandt, E.M., Yu, C.L., Barrick, J.E., Subramanian, M.V. (2013) Caffeine Junkie: an unprecedented GST-dependent oxygenase required for caffeine degradation by P. putida CBB5. J. Bacteriol. 195:3933-3939. «PubMed» | ||
| 40. | Quandt, E.M., Hammerling, M.J., Summers, R.M., Otoupal, P.B., Slater, B., Alnahhas, R.N., Dasgupta, A., Bachman, J.L., Subramanian, M.V., Barrick, J.E. (2013) Decaffeination and measurement of caffeine content by addicted Escherichia coli with a refactored N-demethylation operon from Pseudomonas putida CBB5. ACS Synth. Biol. 2:301-307. «PubMed» | ||
| 39. | Han, P., Niestemski, L.R., Barrick, J.E., Deem, M.W. (2013) Physical model of the immune response of bacteria against bacteriophage through the adaptive CRISPR-Cas immune system. Phys. Biol. 10:025004. «PubMed» | ||
| 38. | Wielgoss, S.*, Barrick, J.E.*,Tenaillon, O.*, Wiser, M.J., Dittmar, W.J., Cruveiller, S., Chane-Woon-Ming, B., Médigue, C., Lenski, R.E., Schneider, D. (2013) Mutation rate dynamics in a bacterial population reflect tension between adaptation and genetic load. Proc. Natl. Acad. Sci. U.S.A. 110:222-227. DOI: 10.1073/pnas.1219574110 «PubMed» | ||
| 2012 | |||
| 37. | Blount, Z.D., Barrick, J.E., Davidson, C.J., Lenski, R.E. (2012) Genomic analysis of a key innovation in an experimental E. coli population. Nature 489:513-518. «PubMed» | ||
| 36. | Jerome, J.P., Klahn, B., Bell, J.A., Barrick, J.E., Brown, C.T., Mansfield, L.S. (2012) Draft genome sequences of two Campylobacter jejuni clinical isolates, NW and D2600. J.Bact. 194:5707-5708. | ||
| 35. | Reba, A., Meyer, A.G., Barrick, J.E. (2012) Computational tests of a thermal cycling strategy to isolate more complex functional nucleic acid motifs from random sequence pools by in vitro selection. In: C. Adami et al. (eds.). Artificial Life XIII: Proceedings of the Thirteenth International Conference on the Synthesis and Simulation of Living Systems. pp 473-480. Cambridge, MA: MIT Press. Awarded Best Synthetic Biology Paper. «Full Text» | ||
| 34. | Kholmanov, I.N., Stoller, M.D., Edgeworth, J., Lee, W.H., Li, H., Lee, J., Barnhart, C., Potts, J.R., Piner, R., Akinwande, D., Barrick, J.E., Ruoff, R.S. (2012) Nanostructured hybrid transparent conductive films with antibacterial properties. ACS Nano 6:5157-5163. «PubMed» | ||
| 33. | Wolf, L.N., Barrick, J.E. (2012) Tracking winners and losers in E. coli evolution experiments. Microbe Magazine 2 (3):124-128. «Article» | ||
| 32. | Meyer, J.R., Dobias, D.T., Weitz, J.S., Barrick, J.E., Quick, R.T., Lenski, R.E. (2012) Repeatability and contingency in the evolution of a key innovation in phage lambda. Science 335:428-432. «PubMed» | ||
| 2011 | |||
| 31. | Nahku, R., Peebo, K., Valgepea, K., Barrick, J.E., Adamberg, K., Vilu, R. (2011) Stock culture heterogeneity rather than new mutational variation complicates short-term cell physiology studies of Escherichia coli K-12 MG1655 in continuous culture. Microbiology 157:2604-2610. «PubMed» | ||
| 30. | Wielgoss, S., Barrick, J.E., Tenaillon, O., Cruveiller, S., Chane-Woon-Ming, B., Médigue, C., Lenski, R.E., Schneider, D. (2011) Mutation rate inferred from synonymous substitutions in a long-term evolution experiment with Escherichia coli. G3: Genes, Genomes, Genetics 1:183-186. «PubMed» |
||
| 29. | Woods, R.J.*, Barrick, J.E.*^, Cooper, T.F., Shrestha, U., Kauth, M.R., Lenski, R.E.^ (2011) Second-order selection for evolvability in a large Escherichia coli population. Science 331:1433-1436. «PubMed» «Press Links» «Science Podcast» «Faculty of 1000» «Erratum» | ||
| 28. | Jerome, J.P., Bell, J.A., Plovanich-Jones, A.E., Barrick, J.E., Brown, C.T., Mansfield, L.S. (2011) Standing genetic variation in contingency loci drives the rapid adaptation of Campylobacter jejuni to a novel host. PLoS ONE 6:e16399. «PubMed» «Blog Article» | ||
| 2010 and earlier | |||
| 27. | Barrick, J.E., Kauth, M.R., Strelioff, C.C., and Lenski, R.E. (2010) Escherichia coli rpoB mutants have increased evolvability in proportion to their fitness defects. Mol. Biol. Evol. 27:1338-1347. «PubMed» | ||
| 26. | Barrick, J.E.*, Yu, D.S.*, Yoon, S.H., Jeong, H, Oh, T.K., Schneider, D., Lenski, R.E., and Kim, J.F. (2009) Genome evolution and adaptation in a long-term experiment with Escherichia coli. Nature. 461:1243-1247. «PubMed»«Faculty of 1000» |
||
| 25. | Barrick, J.E. and Lenski, R.E. (2009) Genome-wide mutational diversity in an evolving population of Escherichia coli. Cold Spring Harbor Symp. Quant. Biol. 74:119-129. «PubMed» | ||
| 24. | Barrick, J.E. (2009) Predicting riboswitch regulation on a genomic scale. In Riboswitches: Methods and Protocols. (ed. A. Serganov), pp. 1-13. Humana Press, New York. «PubMed» |
 |
|
| 23. | Regulski, E.E., Moy, R.H., Weinberg, Z., Barrick, J.E., Yao, Z., Ruzzo, W.L., and Breaker, R.R. (2008) A widespread riboswitch candidate that controls bacterial genes involved in molybdenum cofactor and tungsten cofactor metabolism. Mol. Microbiol. 68:918-932. «PubMed» | ||
| 22. | Weinberg, Z., Regulski, E.E., Hammond, M.C., Barrick, J.E., Yao, Z., Ruzzo, W.L., and Breaker, R.R. (2008) The aptamer core of SAM-IV riboswitches mimics the ligand-binding site of SAM-I riboswitches. RNA 14:822-828. «PubMed» | ||
| 21. | Barrick, J.E. and Breaker R.R. (2007) The distributions, mechanisms, and structures of metabolite-binding riboswitches. Genome Biology 8:R239. «PubMed» | ||
| 20. | Weinberg, Z., Barrick, J.E., Yao, Z., Roth, A., Kim, J.N., Gore, J., Wang, J.X., Lee, E.R., Block, K.F., Sudarsan, N, Neph, S., Tompa, M., Ruzzo, W.L., Breaker, R.R. (2007) Identification of 22 candidate structured RNAs in bacteria using the CMfinder comparative genomics pipeline. Nucleic Acids Res. 35:4809-4819 «PubMed» | ||
| 19. | Yao, Z., Barrick, J.E., Weinberg, Z., Neph, S., Breaker, R.R., Tompa, M., Ruzzo, W.L. (2007) A computational pipeline for high-throughput discovery of cis-regulatory noncoding RNA in prokaryotes. PLOS Comput. Biol. 3:e126. «PubMed» | ||
| 18. | Roth, A., Winkler, W.C., Regulski, E.E., Lee, B.W., Lim, J., Jona, I., Barrick, J.E., Ritwik, A., Kim, J.N., Welz, R., Iwata-Reuyl D., Breaker R.R. (2007) A riboswitch selective for the queuosine precursor preQ1 contains an unusually small aptamer domain. Nat. Struct. Mol. Biol. 14 (4):308-317. «PubMed» | ||
| 17. | Barrick, J.E. and Breaker R.R. (2007) The Power of Riboswitches. Scientific American 296 (1):50-57. «PubMed» | ||
| 16. | Lenski, R.E., Barrick, J.E., and Ofria, C. (2006) Balancing Robustness and Evolvability. PLoS Biology 4:e428. «PubMed» | ||
| 15. | Puerta-Fernandez, E., Barrick, J.E., Roth, A., and Breaker, R.R. (2006) Identification of a new, non-coding RNA in extremophilic eubacteria. Proc. Natl. Acad. Sci U.S.A 103:19490-19495. «PubMed» | ||
| 14. | Sudarsan N., Hammond M.C., Block K.F., Welz R., Barrick J.E., Roth A., Breaker R.R. (2006) Tandem riboswitch architectures exhibit complex gene control functions. Science 314:300-304. «PubMed» | ||
| 13. | Corbino, K.A., Barrick, J.E., Lim, J., Welz, R., Tucker, B.J., Puskarz, I., Mandal, M., Rudnick, N.D., and Breaker, R.R. (2005) Evidence for a second class of S-adenosylmethionine riboswitches and other regulatory RNA motifs in alpha-proteobacteria. Genome Biol. 6: R70. «PubMed» | ||
| 12. | Barrick, J.E., Sudarsan, N., Weinberg, Z., Ruzzo, W.L., and Breaker, R.R. (2005) 6S RNA is a widespread regulator of eubacterial RNA polymerase that resembles an open promoter. RNA 11:774-784. «PubMed» | ||
| 11. | Mandal, M., Lee, M., Barrick, J.E., Weinberg, Z., Emilsson, G.M., Ruzzo, W.L., and Breaker, R.R. (2004) A glycine-dependent riboswitch that uses cooperative binding to control gene expression. Science 306:275-279. «PubMed» | ||
| 10. | Barrick, J.E., Corbino, K.A., Winkler, W.C., Nahvi, A., Mandal, M., Collins, J., Lee, M., Roth, A., Sudarsan, N., Jona, I., Wickiser, J.K., and Breaker, R.R. (2004) New RNA motifs suggest an expanded scope for riboswitches in bacterial genetic control. Proc. Natl. Acad. Sci. U.S.A. 101:6421-6426. «PubMed» | ||
| 9. | Nahvi, A., Barrick, J.E., and Breaker, R.R. (2004) Coenzyme B12 riboswitches are widespread genetic control elements in prokaryotes. Nucleic Acids Res. 32:143-150. «PubMed» | ||
| 8. | Mandal, M., Boese, B., Barrick, J.E., Winkler, W.C., and Breaker, R.R. (2003) Riboswitches control fundamental biochemical pathways in Bacillus subtilis and other bacteria. Cell 113:577-586. «PubMed» | ||
| 7. | Sudarsan, N., Barrick, J.E., and Breaker, R.R. (2003) Metabolite-binding RNA domains are present in the genes of eukaryotes. RNA 9:644-647. «PubMed» | ||
| 6. | Winkler, W.C., Nahvi, A., Sudarsan, N., Barrick, J.E., and Breaker, R.R. (2003) An mRNA structure that controls gene expression by binding S-adenosylmethionine. Nat. Struct. Biol. 10:701-707. «PubMed» | ||
| 5. | Barrick, J.E., and Roberts, R.W. (2003) Achieving specificity in selected and wild-type N peptide–RNA complexes: The importance of discrimination against noncognate RNA targets. Biochemistry 42:12998-13007. «PubMed» | ||
| 4. | Barrick, J.E., and Roberts, R.W. (2002) Sequence analysis of an artificial family of RNA-binding peptides. Protein Sci. 11:2688-2696. «PubMed» | ||
| 3. | Barrick, J.E., Takahashi, T.T., Ren, J.S., Xia, T.B., and Roberts, R.W. (2001) Large libraries reveal diverse solutions to an RNA recognition problem. Proc. Natl. Acad. Sci. U.S.A. 98:12374-12378. «PubMed» | ||
| 2. | Barrick, J.E., Takahashi, T.T., Balakin, A., and Roberts, R.W. (2001) Selection of RNA-binding peptides using mRNA-peptide fusions. Methods 23:287-293. «PubMed» | ||
| 1. | Liu, R.H., Barrick, J.E., Szostak, J.W., and Roberts, R.W. (2000) Optimized synthesis of RNA-protein fusions for in vitro protein selection. Methods Enzymol. 318:268-293. «PubMed» | ||
Ph.D Thesis
| Barrick JE. (2006) Advisor: Ronald R. Breaker. Discovering and defining metabolite-binding riboswitches and other structured regulatory RNA motifs in bacteria. «PDF» |  |
Barrick Lab > PublicationList