Barrick Lab :: Research

Engineering Insect Symbionts
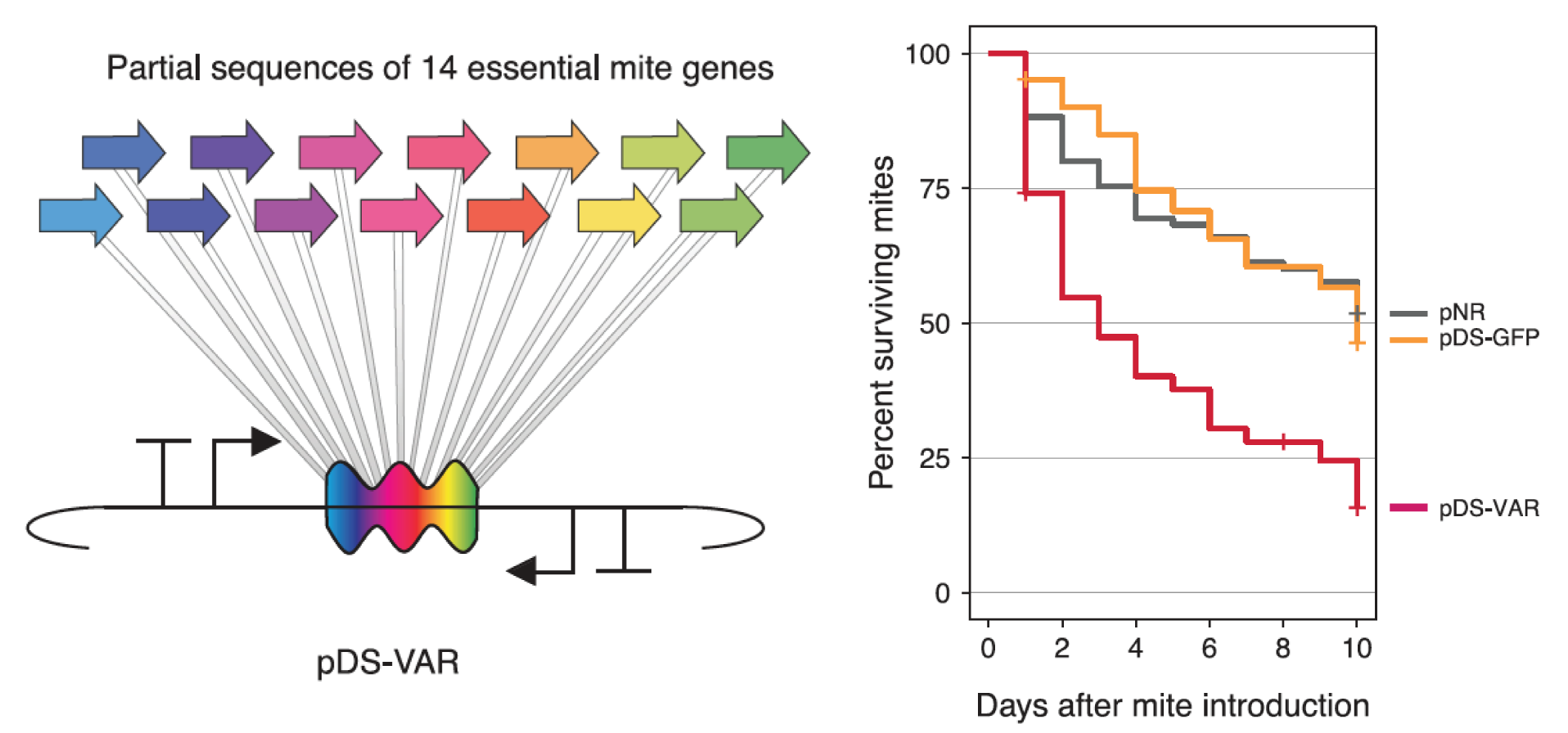
Protection against mites
We created the bee microbiome toolkit (BTK) that can be used to engineer bacteria found in various insects and their gut microbiomes, as demonstrated in honey bees (Apis mellifera) and bumble bees (Bombus species). These tools were used to engineer a native gut bacterium, Snodgrassella alvi, to produce dsRNA targeting two causes of colony collapse: Varroa mites and deformed wing virus. Engineered S. alvi was shown to kill the mites and improve bee health. This symbiont-mediated RNAi approach is helpful to protect insects that are beneficial to human health, but this approach could also be applied to pest species to improve food security or study the biology of the relationship between host and symbiont.
Representative Publications
- Leonard et al. (2020) Science PMID: 32001655
- Leonard et al. (2018) ACS Synthetic Biol. PMID: 29608282
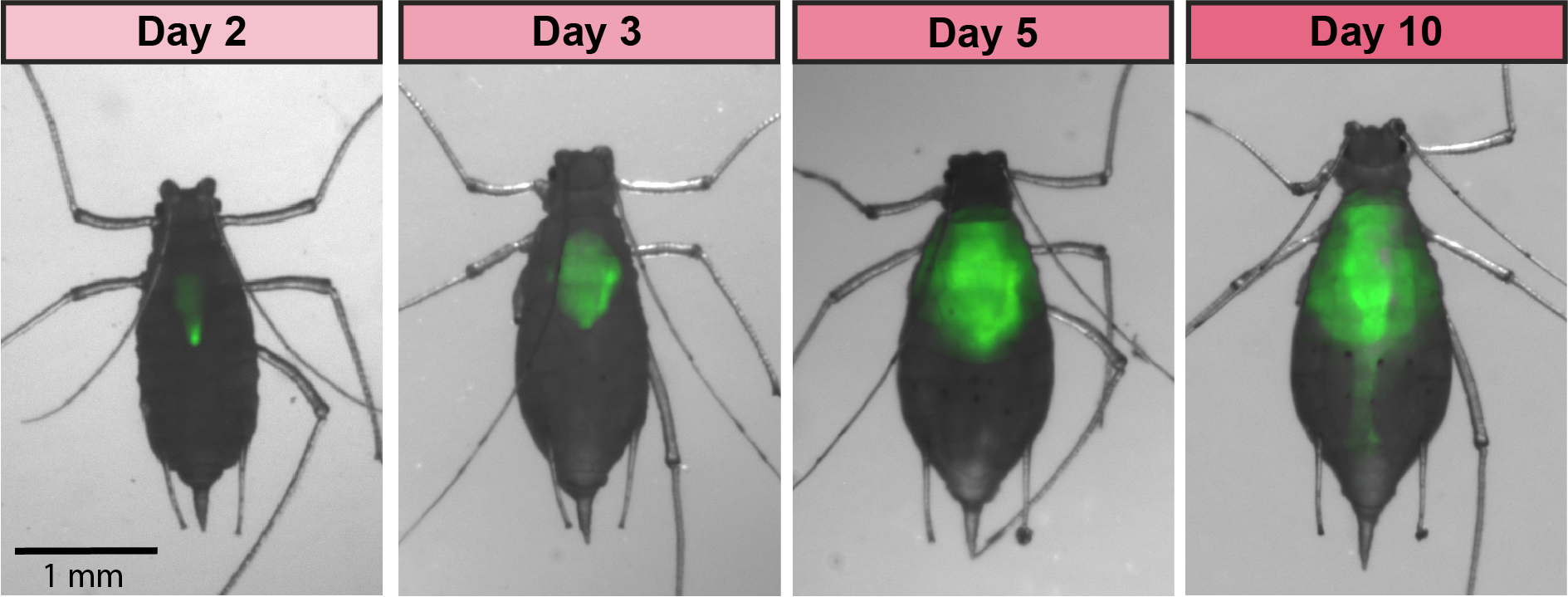
Timelapse of an aphid colonized with GFP-expressing S. symbiotica CWBI-2.3T
In aphids, we are engineering strains of the symbiotic species Serratia symbiotica, cultured from the aphid gut, to enable improved understanding of its relationship with aphids and the development of new tools for food security. Genetic tools for the study and manipulation of aphids have been lacking, but engineering a symbiont for “paratransgenesis” can serve as a useful alternative. We engineered the strain, recolonized aphids with it, and used it to induce aphid mortality. We also showed that cultured S. symbiotica strains are capable of maternal transmission within aphids, suggesting that they possess a latent capacity for a long-term symbiotic relationship with their hosts.
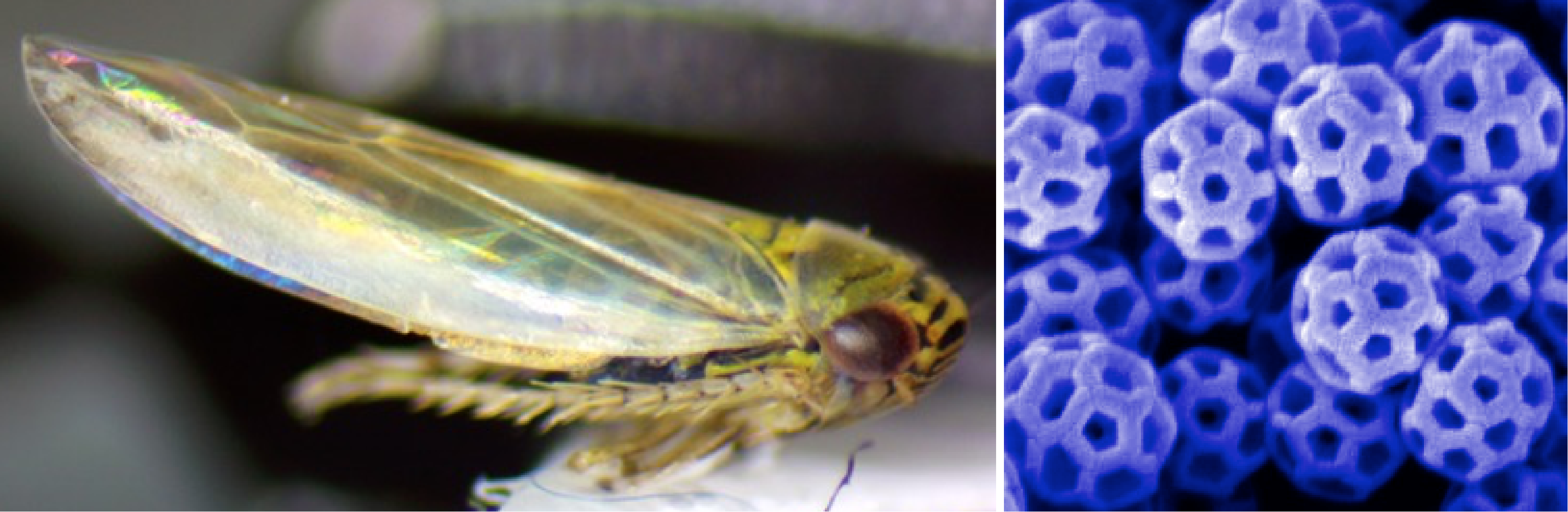
Aster yellow leafhopper (Macrosteles quadrilineatus) and an SEM image of their brochosomes
A newer project in the lab involves another pest insect known as the leafhopper. Leafhoppers have unique nanostructures, called brochosomes, that they anoint onto their wings and sometimes eggs. Brochosomes have special properties, including superhydrophobicity and omnidirectional antireflectivity. We are collecting leafhoppers across Texas with the goal of identifying and characterizing natural variation in brochosome structure across species, ecoregions and seasons. We are working towards engineering leafhopper symbionts to control the production of brochosomes and enhancing their properties for the creation of novel biomaterials using RNAi.
Funding: DARPA BRICS, DARPA AEPHID, MURI ARO
Preventing Evolutionary Failure in Synthetic Biology
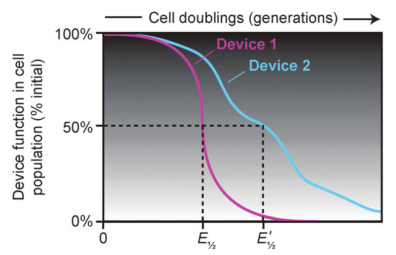
Evolutionary half-lives of biological devices
- Jack et al. (2015) ACS Synthetic Biol. PMID:26096262
- Renda et al. (2014) Mol. Biosyst. PMID:24556867
Dynamics of Microbial Genome Evolution
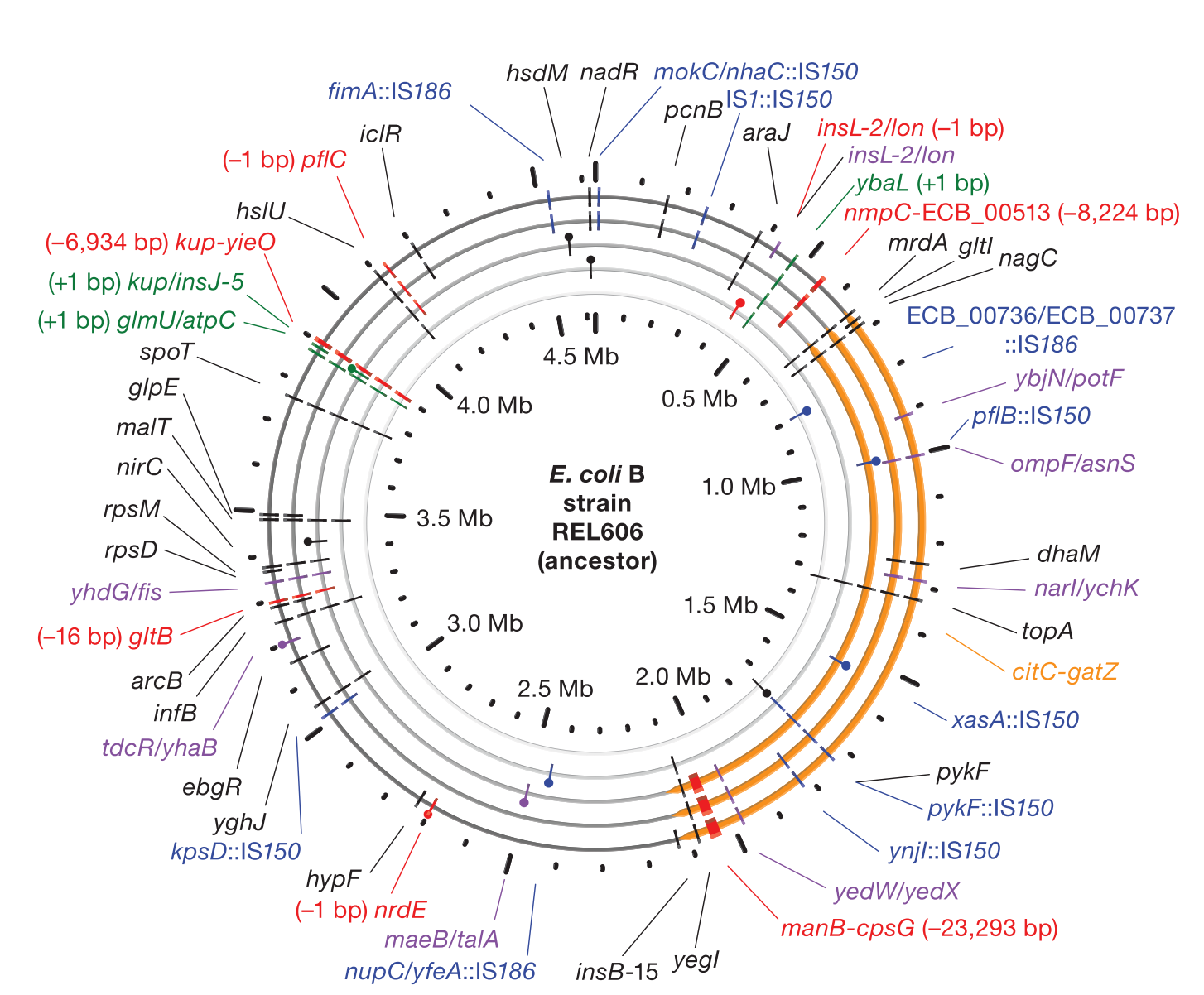
Accumulation of mutations in population Ara-1 of the LTEE over 20,000 generations of evolution
- Deatherage et al. (2017) Proc. Natl. Acad. Sci. PMID: 28202733
- Tenaillon et al. (2016) Nature PMID: 27479321
- Deatherage and Barrick (2014) Methods Mol. Biol PMID:24838886
- Barrick and Lenski (2014) Nat. Rev. Genet. PMID:24166031
Evolution and Engineering of Naturally Transformable Bacteria
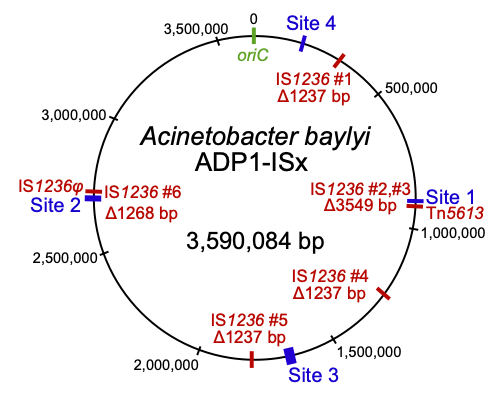
Insertion sequences (red) deleted in the transposon-free strain Acinetobacter baylyi ADP1-ISx
- Suárez et al. (2020) Nucleic Acids Res. PMID:32232367
- Suárez et al. (2017) Appl. Env. Microbiol. PMID:28667117
- Renda et al. (2015) J. Bacteriol. PMID:25512307
Evolution and Engineering of Bacteriophages
- Peng Geng
- Cameron Roots
- UT Austin iGEM
- In colaboration with the Wilke Lab
- Hammerling et al. (2014) Nature Chemical Biology. PMID:24487692
Barrick Lab > ResearchInterests
Contributors to this topic  JeffreyBarrick, KateElston, SarahBialik, IsaacGifford, AlexaMorton, JuliePerreau, GabrielSuarez, CameronRoots
JeffreyBarrick, KateElston, SarahBialik, IsaacGifford, AlexaMorton, JuliePerreau, GabrielSuarez, CameronRoots
Topic revision: r44 - 2020-08-10 - 17:31:19 - Main.CameronRoots

 Mol Biosciences
Mol Biosciences The LTEE
The LTEE iGEM team
iGEM team NGS course
NGS course