Dear internet,
We don't distribute this plasmid. If you want to purify your own enzymes to save money, here are some options to investigate:
|
Protocol for harvesting Pfu-Sso7d polymerase
This protocol is for expressing and purifying the Pfu-Sso7d polymerase from
E. coli [1]. A variant of this protein with an additional 65 amino acid changes is sold as Phusion polymerase by
New England Biolabs. This Pfu variant has the Sso7d processivity-enhancing domain attached that increases its speed and processivity. It generates blunt-end DNA products and typically you use higher annealing temperatures than when using Taq polymerase. The reported differences in fidelity between Phusion and Pfu-Sso7d compared to Taq are 84× and 32×, respectively. See the
NEB website for a description of other key polymerase characteristics.
 Download 6his-Pfu-Sso7d-pET28 expression plasmid sequence (GenBank format)
Download 6his-Pfu-Sso7d-pET28 expression plasmid sequence (GenBank format)
Materials needed
- Glycerol stock of EQ458 E. coli cells
The strain used is named EQ458. It is located in common species box; this is a Rosetta 2 (DE3) E. coli strain containing 6his-Pfu-Sso7d-pET28 plasmid. The plasmid is Kanr and the strain itself is Camr . The frozen stock is overnight growth of a single colony.
- LB medium (recipe)
- Chloramphenicol stock (recipe)
- Kanamycin stock (recipe])
- Refrigerated centrifuge
- Spectrophotometer and cuvettes
- French press (Ex: Georgiou lab, MBB 3.310, ask before using!)
- IPTG (100 mM) stock. Dissolve 2.38 g IPTG in 100 mL deionized water. Filter sterilize and store at –20°C.
- Disposable plastic columns. ThermoSci, cat #29922
- Ni-NTA agarose resin. Qiagen, cat #30210, 25 ml
- Slide-A-Lyzer, 10k dialysis cassette G2. ThermoSci, cat# 87730
Lysis Buffer
- 50 mM NaH2PO4
- 300 mM NaCl
- 10mM Imidazole
- Adjust pH to 8.0 using NaOH
Wash Buffer
- 50 mM NaH2PO4
- 300 mM NaCl
- 40mM Imidazole
- Adjust pH to 8.0 using NaOH
Elution Buffer
- 50 mM NaH2PO4
- 300 mM NaCl
- 250 mM Imidazole
- Adjust pH to 8.0 using NaOH
Polymerase storage buffer Make 3-4 Liters
- 50% Glycerol
- 100 mM Tris/HCl pH 8.0
- 0.2 mM EDTA
- 0.2% NP-40; nonionic detergent
- 0.2% Tween20
- 2 mM DTT (add immediately before using for storage)
 IMPORTANT
IMPORTANT: Add fresh DTT immediately before using a batch of storage buffer to store newly purified enzyme. Your fresh DTT should be newly dissolved from powder or from a 1 M stock that you store frozen at –20°C to avoid oxidation. We have observed rapid loss of function of enzyme when enzyme is diluted in old storage buffer that had DTT added and was then stored at room temperature.
Protein Expression
Scaled for 2×500 mL cultures
Day –1: Revive and Isolate Colony
- Streak LB plate supplemented with Kan and Cam from frozen stock of EQ458. Growth plate overnight at 37°C.
Day –2: Precondition
- Select single colony from O/N streak plate and inoculate 1.5 mL of LB broth supplemented with Kan and Cam. Grow overnight at 37 C shaking at 250 rpm.
Day –1: Induce
- Use 500 µL of overnight culture to inoculate 500 mL of supplemented LB broth (in 2 L flask), grow as before for ~ 3-4 hours until an OD600 of between 0.4 and 0.6 is reached.
- Induce the cultures to express proteins by adding IPTG at a final concentration of 0.5 mM (2.5 mL per 500 mL) followed by overnight growth at 18°C, 250 rpm.
Day 0: Harvest
- Collect cells by centrifugation. Conditions as follows: 4°C, at 10,000 x g for 15 mins.
- Resuspend each cell pellet with 3 mL of lysis buffer and combine tubes together, mix well using pipette.
- French press; use the full cell holding (10 mL - 35 mL) and 1500 psi pressure.
- Collect and reintroduce into French press 1x.
- Heat denature at 70°C for about 15 mins.
- Spin down, 10,000 x g for 30 mins.
- Syringe filter the supernatant (0.22 µm filter).
- Proceed to IMAC purifications.
Immobilized metal ion affinity chromatography (IMAC) purification
Note: Save portions at each step for protein gel
- Prepare a 1 mL Ni-NTA resin column.
- Saturate column with 5× the column volume (so 5 mL) of lysis buffer. Repeat this step twice.
- Bind with 1:1 lysis buffer:SN sample.
- Wash with 1× column volume of lysis buffer.
- Wash with 5× (column volume) of wash buffer.
- Elute with 3 mL of elution buffer and collect all 3 mL of elution.
 NOTE
NOTE: This purification protocol does not perfectly purify the polymerase from all other
E. coli proteins. Expect your protein sample to still be heterogeneous if you run it on a gel. Don't worry, it will have sufficient purity for DNA amplification. The heating step at 70°C during harvesting helps precipitate many
E. coli proteins.
Dialysis:
- Place dialysis cassette into storage buffer for 2 mins.
- Remove top and load dialysis cassette with enzyme sample using a pipette or syringe.
- Squeeze the membrane to remove excess air.
- Replace top and place in beaker with 500 mL or 1 L of storage buffer. This should be done in the cold room on a stir plate.
- Allow to sit for 2 to 4 hours.
- Remove cassette and place in beaker with fresh storage buffer. Allow to sit overnight.
- If cassette has swollen, use syringe to remove some of the sample.
- Open top of cassette and remove sample.
- Store your newly purified enzyme at –20°C.
Assay purified polymerase activity by PCR
- IMPORTANT: You probably want to use commercial Phusion buffer (NEB Cat #B0518S) for your reactions. It is a proprietary formulation that gives MUCH better enzyme performance.
- The template for this assay is the 6his-pfu-sso7d-pET28 plasmid encoding the polymerase.
- To estimate the activity, create a dilution series of purified polymerase in water ranging from 1:200 to 1:10, and compare to NEB Phusion or a similar polymerase.
- NEB stock is viscous; for an accurate comparison to the purified polymerase, ensure you are pipetting sufficient volumes to maintain accuracy.
- After measuring the activity of your main stock, be sure to use storage buffer that has fresh DTT added to it for making any working dilutions of your original high concentration stock that will be stored for long periods of time at –20°C.
Primer sequences (position with reference to sequence in file above)
| Name |
Position |
Tm |
5' - 3' |
Amplicon size w/ R1 (bp) |
| Phusion F1 |
672-695 |
66.4 |
agttccataggatggcaagatcc |
4044 |
| Phusion F2 |
2748-2770 |
66.5 |
tgataccgatgaaacgagagagg |
1968 |
| Phusion F3 |
3759-3779 |
66.4 |
gagctgtcttcggtatcgtcg |
957 |
| Phusion F4 |
4169-4190 |
66.7 |
aacattagtgcaggcagcttcc |
547 |
| Phusion R1 |
4694-4716 |
67.8 |
cctaatgcaggagtcgcataagg |
NA |
Reaction Mix
| Reagent |
Volume/ul |
| Water |
12.4 |
| dNTP (10mM) |
0.4 |
| 5× buffer |
4 |
| Primer F (10uM) |
1 |
| Primer R (10uM) |
1 |
| Template (2ng/ul) |
1 |
| Diluted polymerase |
0.2 |
* PCR Conditions* (Denaturation-Annealing-Extension repeated 30x):
| Cycle |
Temperature |
Duration (secs) |
| Initial denaturation |
98 |
30s |
| Repeat Cycle 30× |
| Denaturation |
98 |
10s |
| Annealing |
69 |
20s |
| Extension |
72 |
30s / kbp |
| End Cycle |
| Final extension |
72 |
5m |
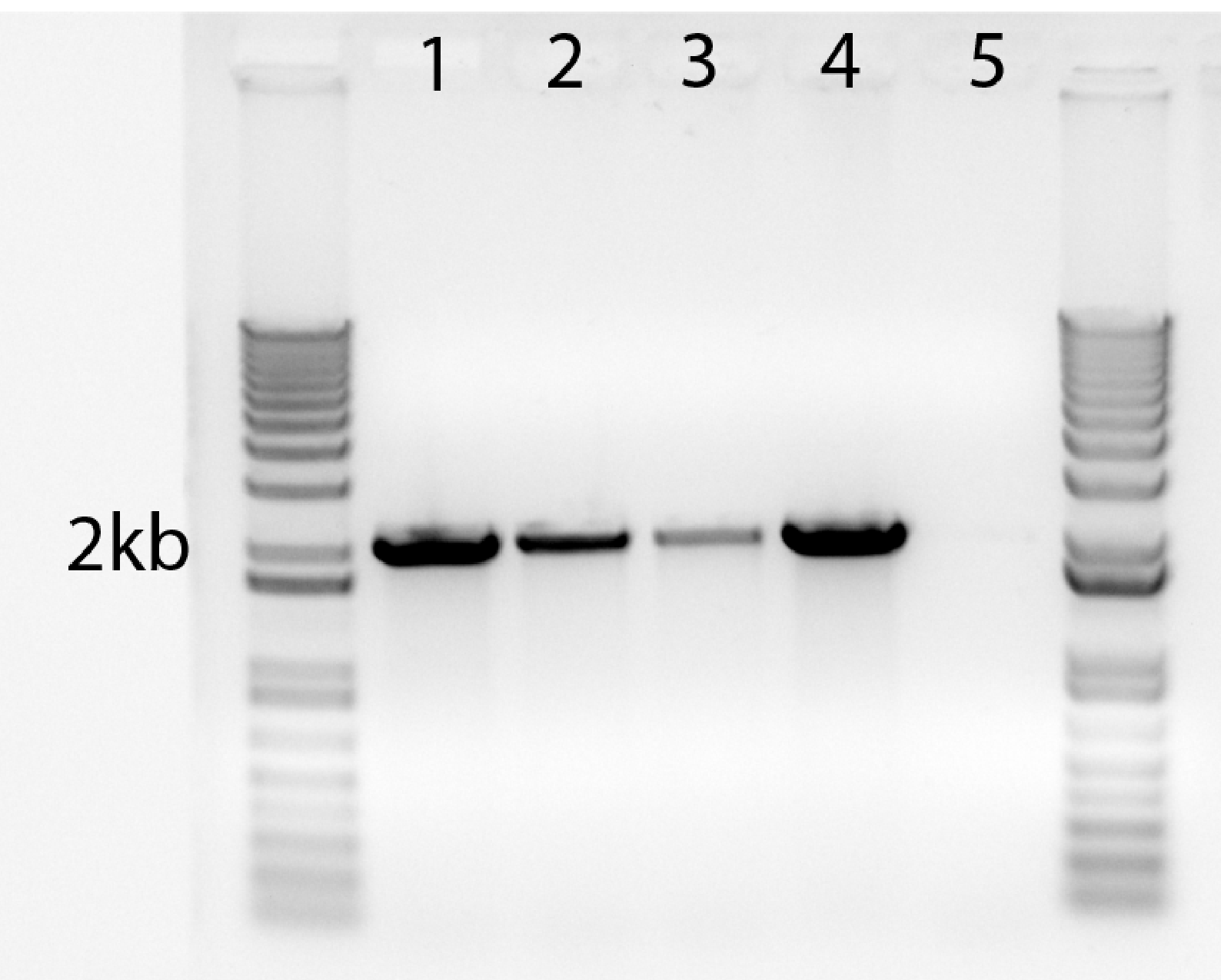
Sample results for 2-kb amplicon
| Lane |
1 |
2 |
3 |
4 |
5 |
| Phusion dilution |
25 |
50 |
100 |
NEB (neat) |
H2O |
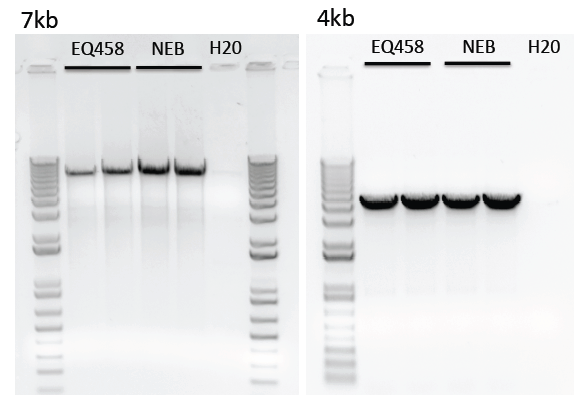
Limitations of purified polymerase
We have noticed reduced amplification of longer transcripts (>4kb) using our purified polymerase compared to the commercial enzyme.
References
- Wang, Y., Prosen, D. E., Mei, L., Sullivan, J. C., Finney, M., Vander Horn, P. B. (2004) A novel strategy to engineer DNA polymerases for enhanced processivity and improved performance in vitro. Nucleic Acids Res. 32: 1197–1207. «PubMedCentral»


Barrick Lab > ProtocolList > ProtocolsReagentRecipes > ProtocolsReagentsPfuSso7d