<<Return to qPCR page
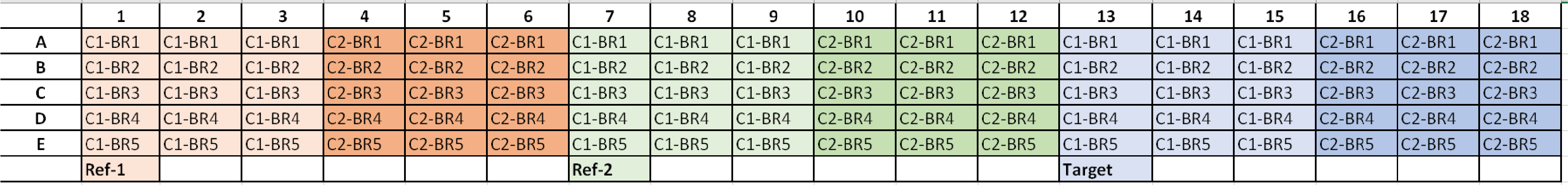 *C1 = Condition 1, C2 = Condition 2, and BR# = Biological Replicate
Conditions
*C1 = Condition 1, C2 = Condition 2, and BR# = Biological Replicate
Conditions
Experimental qPCR Plate Setup and Analysis
Goals- Test hypothesis
 *C1 = Condition 1, C2 = Condition 2, and BR# = Biological Replicate
Conditions
*C1 = Condition 1, C2 = Condition 2, and BR# = Biological Replicate
Conditions
- If you want to be extra safe you should also include a negative control (usually DIW) for each primer pair.
- The cDNA dilution used for the experimental samples will be those determined from the cDNA Concentration qPCR, and you should use at least your best 2 reference genes from the Reference Gene qPCR.
- Technical replicates with a standard deviation below 0.2.
- Outliers; if you have an SD for a technical triplicate higher than 0.2, and there is obviously an outlier, remove it. An example is if you have values of 23.34, 23.35 and 30.22, and the 30.22 amplification trace is clearly poor. If the SD is higher than 0.2 and all three measurements are spread, you must keep all three.
Analysis
I like to perform my qPCR analysis using the pfaffl method, a brief description of which is shown below:The Pfaffl method... is used to calculate relative gene expression data while accounting for differences in primer efficiencies. Unlike the delta-delta Ct method, which assumes primer efficiencies are similar between the gene of interest and the housekeeping gene, the Pfaffl method accounts for any efficiency differences to increase reproducibility.To perform the pfaffl method I've found that the best guide is this website. Dr Steven Bradburn has already done an excellent job of explaining this in a simplified fashion and you should read his guide before starting this procedure! <<Return to qPCR page
-- Top Tips Bio
Barrick Lab > ProtocolList > QPCR > ExperimentalqPCR