<<Return to qPCR page
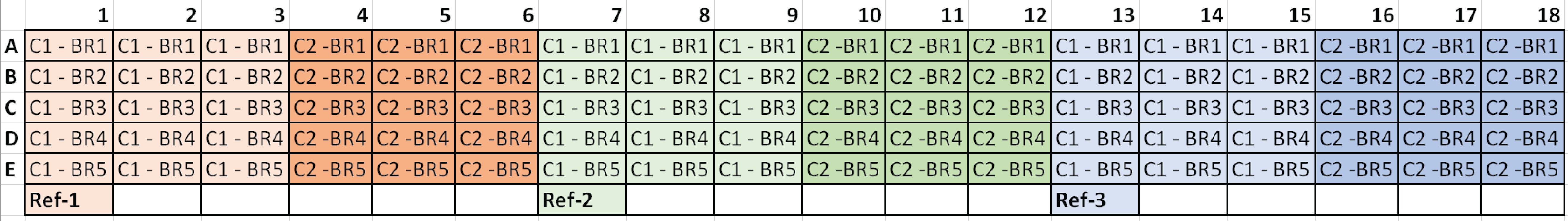 *C1 = Condition 1, C2 = Condition 2, and BR# = Biological Replicate
Template Material
*C1 = Condition 1, C2 = Condition 2, and BR# = Biological Replicate
Template Material
If you are seeing differences between replicates and conditions, it is likely due to technical issues encountered when setting up the qPCR plate. The first few times you run qPCR the results can be messy, and sometimes you have to redo things to get accurate results. That being said, if you're a qPCR pro, or if you keep getting the same results over and over you may have to design primers for a new reference.
<<Return to qPCR page
Reference Gene qPCR
Goals- Determine which of your reference genes you are going to normalize to.
 *C1 = Condition 1, C2 = Condition 2, and BR# = Biological Replicate
Template Material
*C1 = Condition 1, C2 = Condition 2, and BR# = Biological Replicate
Template Material
- Use cDNA dilution determined in cDNA Concentration qPCR
- Technical replicates with a standard deviation below 0.2. This confirms the accuracy of your results.
Analysis
There is no detailed analysis for this step. Look for references that match the "what you are looking for" criteria and select:- At least 2 reference primer sets that show no significant difference between control and experimental conditions. The standard deviation of the Cqs for all biological replicates should be low, preferrably less than 0.5.
If you are seeing differences between replicates and conditions, it is likely due to technical issues encountered when setting up the qPCR plate. The first few times you run qPCR the results can be messy, and sometimes you have to redo things to get accurate results. That being said, if you're a qPCR pro, or if you keep getting the same results over and over you may have to design primers for a new reference.
<<Return to qPCR page
Barrick Lab > ProtocolList > QPCR > RefGeneqPCR