The University of Texas at Austin :: iGEM Team
Interested in joining the UT Austin iGEM Team?
Participating in the iGEM competition is a unique opportunity for undergraduate students to gain research and career skills. iGEM is a science pentathlon! It's part synthetic biology research project, part robotics competition for biology, part entrepreneurial pitch contest, part bioethics course, and part engineering design project. Students are expected to explore their own project ideas, reflect on and explain how their proposed solution to a problem would benefit society, engage in outreach to stakeholders, and operate as a team to complete proof-of-principle research. After working on their research during the spring and summer, teams create a project website and give poster and oral presentations at a Jamboree in mid-fall with >350 teams from all over the world participating.
Applications for the 2022 team will be due Dec. 1st, 2021.
Click here for more information and application instructions!
What is synthetic biology? What is iGEM?
 |
"Synthetic biology is the design and construction of biological devices and systems for useful purposes." –Wikipedia page "The International Genetically Engineered Machine competition (iGEM) is the premiere undergraduate Synthetic Biology competition. Student teams are given a kit of biological parts at the beginning of the summer from the Registry of Standard Biological Parts. Working at their own schools over the summer, they use these parts and new parts of their own design to build biological systems and operate them in living cells." –iGEM Foundation |
2020 Season Project
Bacterial contamination of drinking water is a huge problem worldwide, causing 780,000 deaths on average each year. With PhastPhage, we intend to create a phage-based biosensor for detecting the presence of E. coli contamination in water sources. Using the stoichiometric gene expression simulator Pinetree, we modeled the genome of bacteriophage T7 in order to determine how we could engineer it to produce large amounts of GFP and shorten the time it takes to lyse bacterial cells during an infection cycle. We determined that the most effective way to achieve this was to place a GFP gene, as well as move a holin gene responsible for lysis proteins, into the genome adjacent to gene 10A, which encodes for a major capsid protein and has the highest level of expression of all of T7ís genes. We intend to continue research in the bacteriophage sphere for 2021 with projects utilizing wet lab and computational approaches. You can view our wiki for the 2020 project here and our GitHub repository here.Past UT Austin iGEM Team Projects
UT Austin has a long history of successful participation in iGEM, including:- Implementing bacterial photography (the Coliroid)
- Best Measurement Awards in 2019 and 2012.
- Earning the top Gold Medal designation seven times.
- iGEM team students as co-authors on scientific papers
 2020 iGEM Team Phast-Phage |
 2019 iGEM Team Burden-O-Meter |
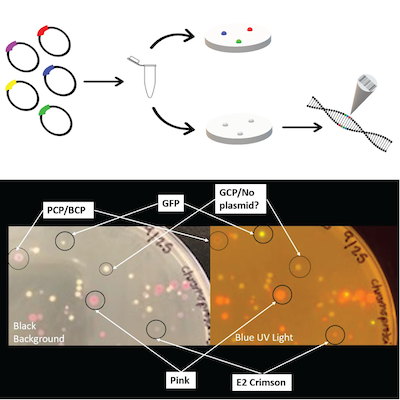 2018 iGEM Team BHR plasmid kit |
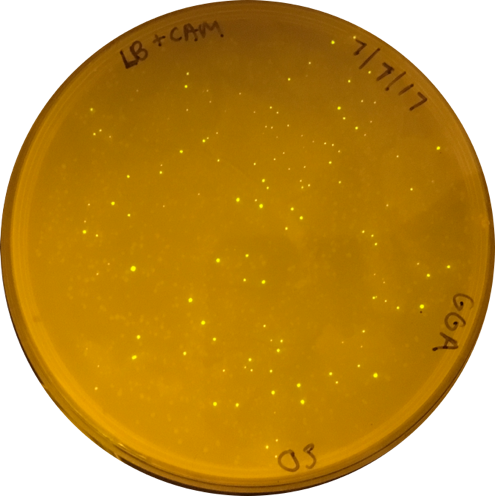 2017 iGEM Team Yo GABA GABA |
 2016 iGEM Team SCOBY Doo |
 2015 iGEM Team Breaking is Bad |
 2014 iGEM Team GC Expansion Pack |
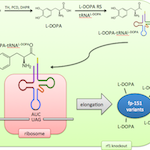 2013 iGEM Team MAPs and Bioscrubber |
 2012 iGEM Team Decaffeination |
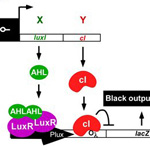 2006 iGEM Team Edge Detector |
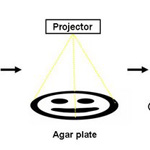 2005 iGEM Team Edge Detector |
 2004 iGEM Team Bacterial Photography |
 |
| Members of the 2019 UT iGEM team at the Giant Jamboree in Boston |
 |
| 2019 UT iGEM team with the Best Measurement Award for their Burden-O-Meter project |
Barrick Lab > UTiGEMTeam
Topic revision: r48 - 2021-02-11 - 18:56:05 - Main.JeffreyBarrick