Handling High Molecular Weight DNA
Introduction
Long-read sequencing, such as Oxford-Nanopore or PacBio sequencing, is becoming increasingly important for genome assembly and genomic research as reads stretching across repeats and mobile genetic elements can preserve structural information about the chromosome unavailable to short-read sequencing. For long-read sequencing high molecular weight DNA (HMW DNA), composed of long fragments generally greater than 50 kb, is necessary. Special care must be taken with these samples to preserve the size of the fragments: routine lab manipulations can break the DNA strands into smaller pieces, reducing the information they contain. As a general rule: minimize pipetting, vortexing, heat stress, and freezing wherever possible when working with HMW DNA extracts. Ultra high molecular weight DNA (UHMW DNA) is even longer than HMW DNA, greater than 250 kb, and may require additional handling considerations. N50 is a measure commonly used to compare the lengths of fragments in HMW samples. The N50 of a sample corresponds to the length of a fragment at which the fragments above and below it have equal total lengths. Or, more simply, if all the fragments were arranged in order from longest to shortest, the N50 would be the length of the fragment in the middle (Figure 1).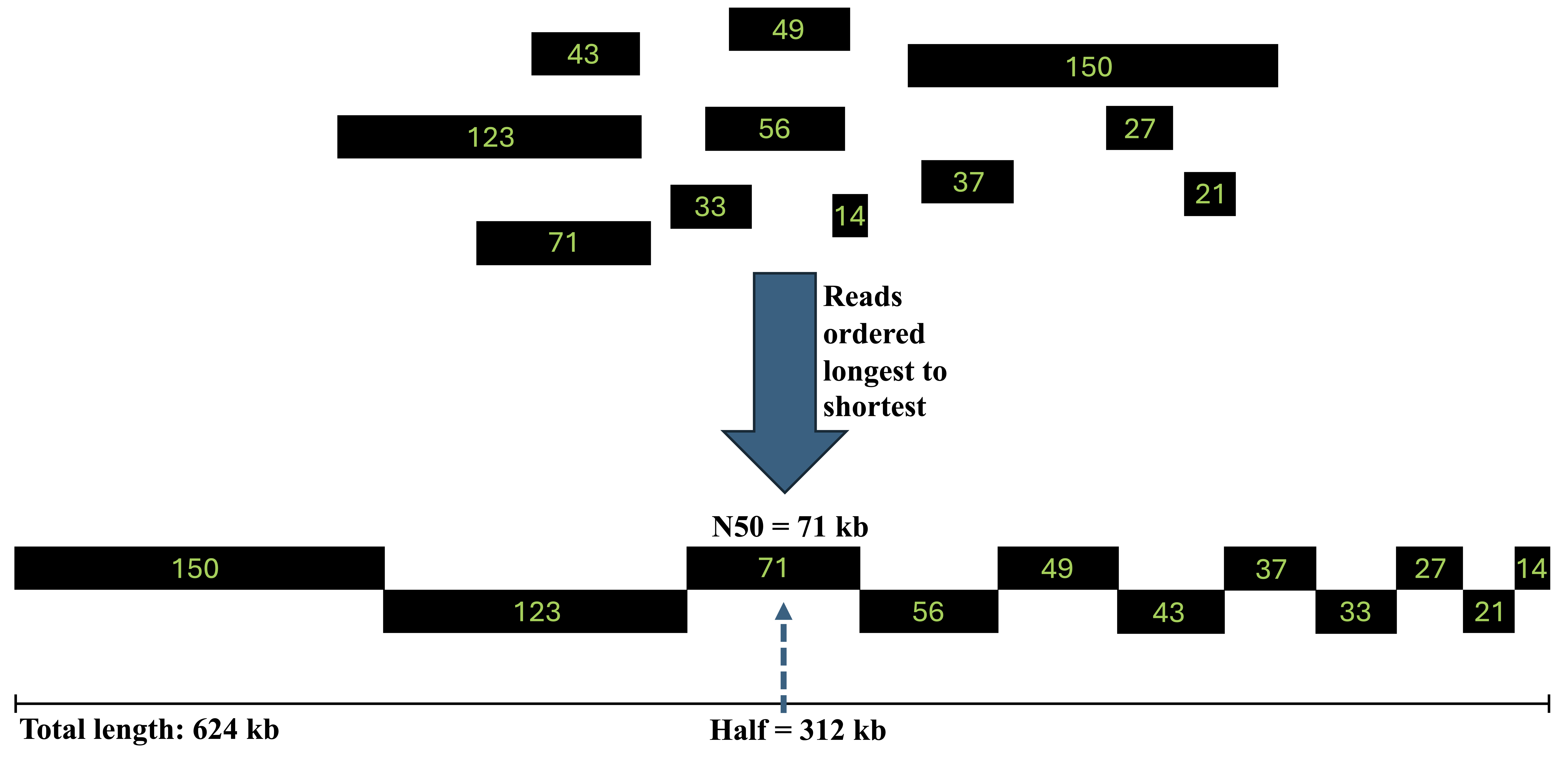
Extraction
Several kits for HMW DNA extraction exist, which utilize magnetic beads or isopropanol precipitation for purification. The Barrick Lab currently uses the Promega Wizard Genomic DNA Purification Kit for HMW DNA extraction. Typical yields from 1 ml of E. coli culture are around 100 μl of extract at 50-200 ng/μl. N50 values when sequenced on an Oxford-Nanopore MinION sequencer are around 10-15 kb with a long-tailed read-length distribution reaching greater than 1 Mb. Note Promega also makes a kit named the Wizard HMW DNA Extraction Kit which is designed for even longer read lengths (up to 500 kb). In our experience the Genomic DNA Purification Kit is sufficient for most general bacterial genome sequencing but the HMW DNA Extraction Kit may be useful for sequencing genomes with large repeat regions.Resuspension and Quantification
After extraction, HMW DNA frequently forms clumps and will not be well distributed within the sample. Resuspension requires incubation of the extract for periods ranging from an hour on a heat block to incubation overnight at room temperature or 4 °C for several days. Gently flicking the tube during resuspension can help disperse the DNA. At high concentrations the resuspended mixture becomes viscous and will pipette slowly. HMW DNA extracts must be resuspended before quantification or the DNA will not be distributed evenly throughout the sample. While fluorescent dye-based quantification methods are typically preferred for quantifying DNA extracts they are less accurate when quantifying HMW DNA. Qubit quantification of HMW DNA can be off by more than 25% when using traditional standards [1]. Using genomic DNA extracts as standards may produce more reliable results.Storage and Shipping
If being used immediately, HMW extracts should be stored at 4 °C. For long-term storage HMW extracts can be frozen at -20 °C. Note that repeatedly freezing and thawing samples can lead to degradation of HMW DNA and lead to a decrease in N50 [2], so this should be used sparingly. HMW DNA can be damaged during shipping. To prevent this it is recommended to ship HMW DNA frozen on dry ice, as frozen samples are less likely to be damaged by jostling during transit [3]. HMW DNA extracts may need to be re-suspended after thawing.MinION Sequencing
Oxford-Nanopore MinION sequencers are lab-scale devices for long-read sequencing. To sequence multiple samples the DNA is barcoded to differentiate reads from each sample before being mixed together to form a library that is then loaded into a flow cell.- There are two main types of barcoding kits: rapid and ligation kits. Rapid kits use a transposase system to insert the barcode into the middle of a fragment of DNA whereas ligation kits use ligase to attach the barcode on the end. Rapid kits are faster and require smaller quantities of DNA, however, the transposase will cleave the DNA during barcoding, decreasing the read length. Thus, if prioritizing longer reads a ligation barcoding kit is preferred.
- Exposure to air can damage the pores in the flow cell, thus special care should be taken when loading the priming mix and library to prevent the injection of air bubbles.
- When loaded properly, the standard output from a single flow cell should be at least 8 Gb of total sequencing [4]. Assembling a bacterial genome de novo will likely require 30-50x coverage (i.e. total sequencing output 30-50 times the size of the strain's genome).
References
[1] https://www.neb.com/en-us/tools-and-resources/usage-guidelines/measuring-analyzing-and-storing-high-molecular-weight-dna-hmw-dna-samples [2] https://nanoporetech.com/document/requirements/dna-stability-f-t [3] https://www.pacb.com/wp-content/uploads/Technical-Note-Preparing-DNA-for-PacBio-HiFi-Sequencing-Extraction-and-Quality-Control.pdf [4] https://nanoporetech.com/document/input-dna-rna-qc Barrick Lab > ProtocolList > HMWDNA