CFU Counts
This is an outline for a general protocol to assess the number of colony forming units in a sample using spot plating. This method cuts down on the number of plates and makes the process better for high throughput experiments. We have used this method to assess bacterial titer in insects and determine conjugation efficiency, but it has plenty of other practical uses.(Optional) Preparation of Sample
This first step is useful to prepare insect samples when calculating bacterial titer or when working with bacteria that are being transferred from one media condition to another (ie following conjugation when samples are transferred from nonselective DAP plates to selective plates). This preparation involves performing washes in a saline solution to remove media/surface contaminants. Insect samples:- Soak your insects in 200 µL 10% Bleach for 1 minute
- Rinse your sample with the same amount of 145 nM Saline (or 1× PBS)
- Resuspend the sample in 200 µL Saline (or PBS) and crush with a micropestle
- Spin down 1 mL culture (or standard amount of pellet resuspended in 1ml Saline) at 3000rpm for 5 minutes (this time can be increased if your bacteria does not pellet)
- Remove the liquid from your sample and resuspend in 1 mL 145 nM Saline
- Repeat the centrifugation and resuspension process for a total of 2 washes
- Resuspend your sample in 200 µL to 1 mL Saline
- This amount will determine how saturated your sample is for the dilution step. If you have a lot of sample, increase the amount of liquid you resuspend in, but make sure that you are consistent in your experiments
Sample Dilution and plating
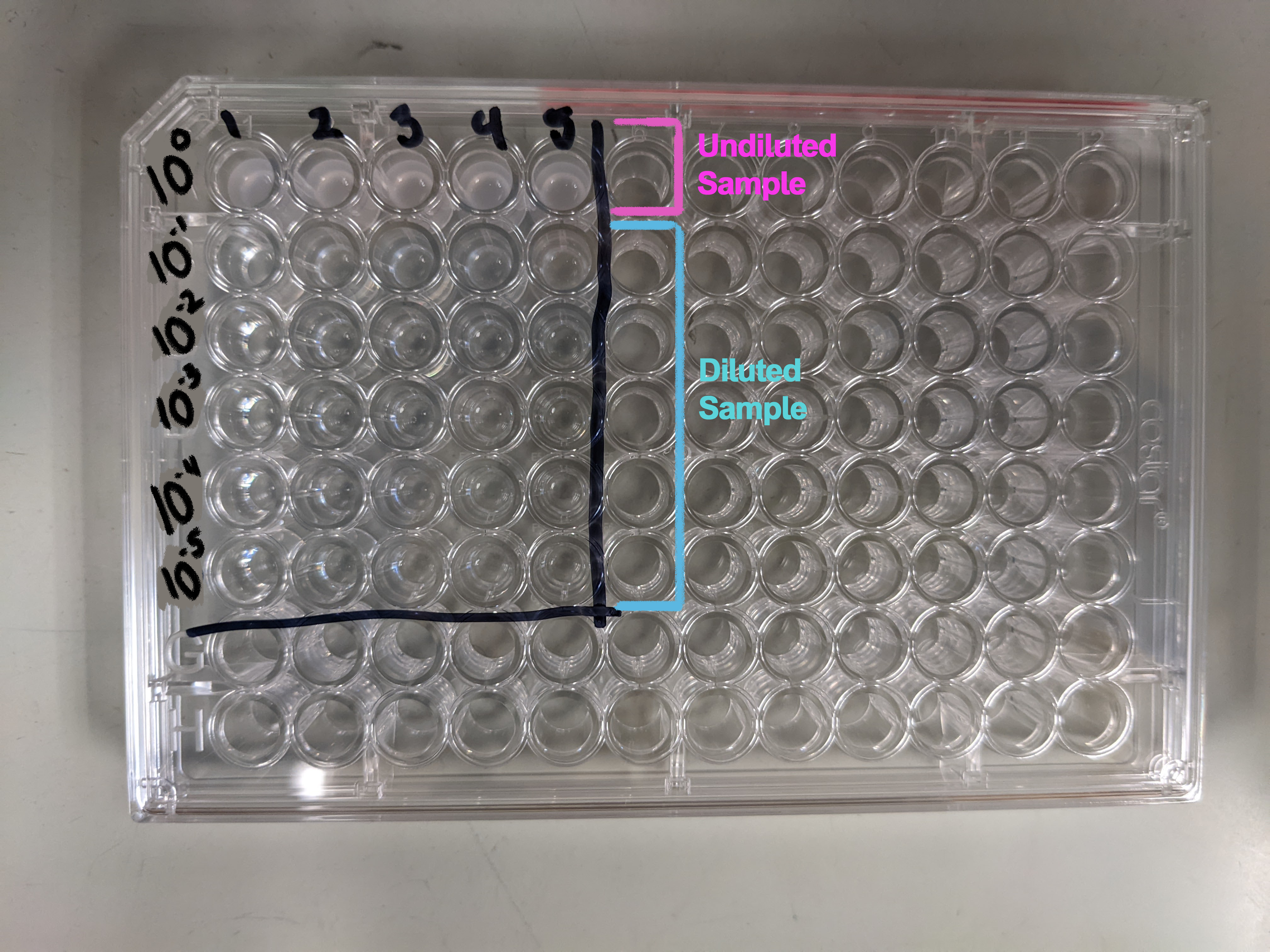
Generally the dilutions you'll want to perform can be done in a 96-well plate. This makes the process compatible with multichannel pipets and the Opentron robot to speed up the process. The image below shows the plate setup with 1:10 dilutions ranging from 100 to 10-5. If, when performing this assay, you find that this range is not useful for your sample you can adjust it by changing the number or scale of the dilutions. What you need: Dilution:- 96-well plate
- 145 nM Saline or 1× PBS (use the same solution as in your sample prep)
- p200 multichannel pipet and tips (decreases contamination risk)
- Reagent reservoir (to use with multichannel)
- p20 multichannel pipet and tips
- DRY agar plates (enough for all your samples plated in triplicate)
- Note: I tend to prefer plates that were poured the day before and left out at room temp to dry overnight. If you are working from plates taken from the deli fridge you should put them in the 37°C incubator a few hours ahead of time to evaporate
- Other note: I recommend drawing a grid onto the bottom of your plates to standardize the plating process and make it easier to see where your samples should go
- Fill each of the wells of the 96-well plate that you will be using with 180 µL Saline/PBS (see image on right)
- Fill the top row of your plate with 200 µL sample
- Using the p200 pipet, perform serial dilutions of 20 µL into each well (20 in 200 = 1:10 dilutions) - Change tips between each transfer!
- Using your p20 pipet - pipet up 5 µL of sample, line it up with the grid on your plate, touch the pipet tips to the plate and slowly depress the plunger to deposit a row of droplets onto the agar
- If your sample spreads into another your plate is not dry enough! Stop what you are doing and let those plates dry
- Plate all your samples in triplicate from the same set of dilutions (see image)
- Allow your spots to dry and then transfer them to the appropriate incubator to grow
CFU counting and calculations
 To count your colonies it is important that they are visible and not running into each other! This can sometimes be challenging when working with 5 µL spots - you'll want to work out the appropriate amount of growth time for whichever bacteria you are testing in this assay.
Counting recommendations:
To count your colonies it is important that they are visible and not running into each other! This can sometimes be challenging when working with 5 µL spots - you'll want to work out the appropriate amount of growth time for whichever bacteria you are testing in this assay.
Counting recommendations: - It's best to count the samples at the lowest dilution where you can easily resolve individual colonies. (see image on right)
- Counting can be facilitated with the colony counter light box or the manual clickers
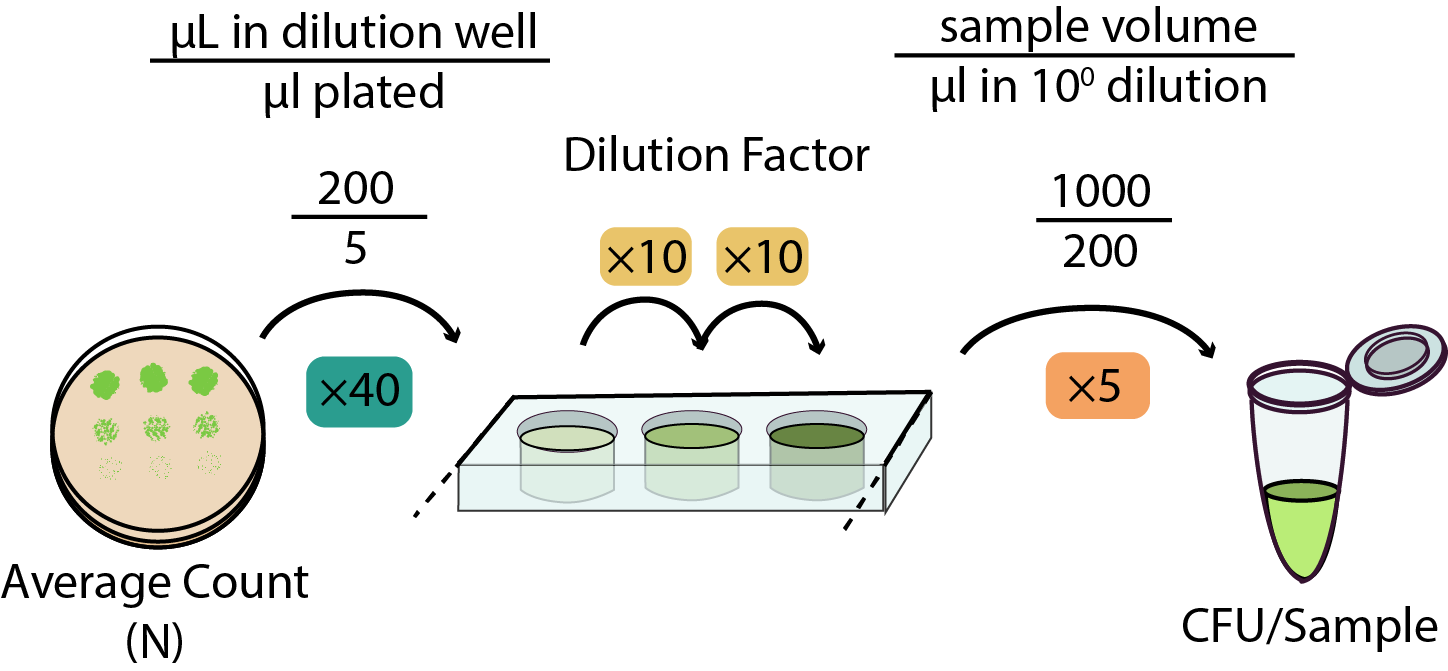
- Make sure you record both the dilution counted and the number of colonies - both numbers are important when calculating the CFU of your sample!
Contributors to this protocol:
-- No permission to view TWiki.UserReportsReferences:
Elston et al., 2021-Use of this protocol to measure bacterial titer in aphids| I | Attachment | History | Action | Size | Date | Who | Comment |
|---|---|---|---|---|---|---|---|
| |
spot_plating_calculator.xlsx | r1 | manage | 10.7 K | 2022-08-05 - 17:19 | UnknownUser |
Barrick Lab > ProtocolList > ProtocolsCFUCounts

