Golden Gate Assembly
This page serves as the main repository for everything Golden Gate Assembly-related. This page links to a number of protocol pages that will help you design DNA sequences for Golden Gate Assembly, learn about the various different Golden Gate Assembly techniques and when we employ each one, and conduct the different types of Golden Gate Assembly reactions. Background & Design:Golden Gate Assembly (GGA) was first described in Engler C, Kandzia R, Marillonnet S (2008) and Engler C, Gruetzner R, Kandzia R, Marillonnet S (2009) as an efficient way to quickly assemble multiple DNA sequences, or parts, into a single plasmid. This molecular cloning method allows a researcher to simultaneously and directionally assemble multiple DNA fragments into a single piece using Type IIs restriction enzymes and T4/T7 DNA ligase. This assembly is performed in vitro. The most commonly used Type IIs enzymes are BsaI, BsmBI and BbsI. Unlike standard Type II restriction enzymes like EcoRI and BamHI, these enzymes cut DNA outside of their recognition sites and therefore can create customized overhangs. Since 256 potential overhang sequences are possible, multiple fragments of DNA can be assembled by using combinations of overhang sequences. Golden Gate Assembly Schematic:
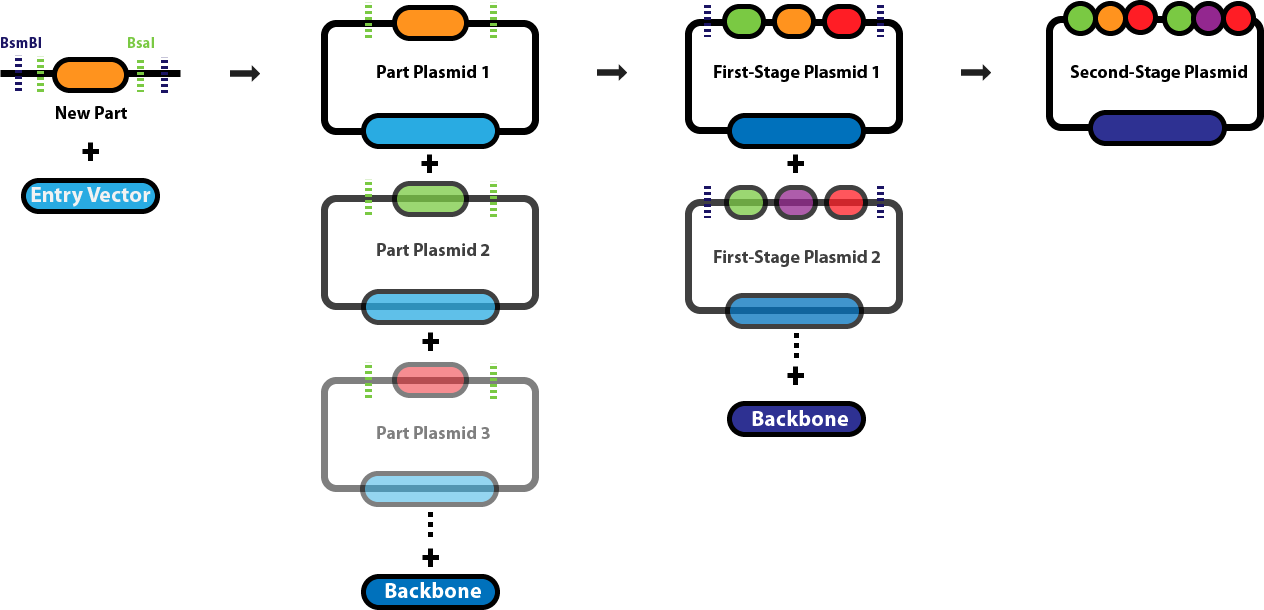
Golden Gate Assembly Links
Golden Gate Assembly - Start to Finish- Design a new part Design a new part with correct part-type overhangs.
- Assemble a new part plasmid Assemble a new part plasmid.
- Build a first-stage plasmid (Transcriptional Unit, BsaI assembly) Build a complete plasmid with a single BsaI reaction.
- Build a second-stage plasmid Assemble multiple first-stage transcriptional unit plasmids.
- Tips, Tricks, and Troubleshooting Running into problems? Check out these potential solutions!
- Bee Toolkit Primer Design: https://spleonard1.shinyapps.io/paRting/
- Reaction Calculation Spreadsheet: BroadHostRange_Reaction_Calculator.xlsx
- NEBeta Tools for the design of high-fidelity Golden Gate Assemblies: https://www.neb.com/research/nebeta-tools
References
- Lee ME, DeLoache WC, Cervantes B, Dueber JE. (2015) A Highly Characterized Yeast Toolkit for Modular, Multipart Assembly. ACS Synth. Biol. 4:975–986. Link
- Engler C, Kandzia R, Marillonnet S. (2008) A one pot, one step, precision cloning method with high throughput capability. PLoS ONE 3:e3647. Link
- "Golden Gate Assembly". New England Biolabs. Retrieved 8 June 2015. Link
- Iverson et al. (2016) CIDAR MoClo: Improved MoClo Assembly Standard and New E. coli Part Library Enable Rapid Combinatorial Design for Synthetic and Traditional Biology. ACS Syn Bio 5 (1), pp 99–103. http://pubs.acs.org/doi/abs/10.1021/acssynbio.5b00124
Contributors
- Peng Geng
- Sean Leonard
- Kate Elston
- Dennis Mishler
- Jeffrey Barrick
-- Main.KateElston - 29 Jan 2018
The excel worksheet for calculating Golden Gate Assembly Reactions can be found in the attachment below.
| I | Attachment | History | Action | Size | Date | Who | Comment |
|---|---|---|---|---|---|---|---|
| |
BroadHostRange_Reaction_Calculator.xlsx | r6 r5 r4 r3 r2 | manage | 17.0 K | 2021-07-09 - 20:54 | KateElston | Golden Gate Reaction Calculator |
| |
golden_gate_diagram.ai | r1 | manage | 389.9 K | 2018-01-30 - 17:16 | KateElston |
Barrick Lab > ProtocolList > GoldenGateAssemblyProtocolsMainPage
Topic revision: r14 - 2021-07-12 - 21:51:28 - Main.KateElston

