Golden Gate Assembly
This page serves as the main repository for everything Golden Gate Assembly-related. This page links to a number of protocol pages that will help you design DNA sequences for Golden Gate Assembly, learn about the various different Golden Gate Assembly techniques and when we employ each one, and conduct the different types of Golden Gate Assembly reactions. Background & Design:Golden Gate Assembly (GGA) was first described in Engler C, Kandzia R, Marillonnet S (2008) and Engler C, Gruetzner R, Kandzia R, Marillonnet S (2009) as an efficient way to quickly assembly multiple DNA sequences, or parts, into a single plasmid. This molecular cloning method allows a researcher to simultaneously and directionally assemble multiple DNA fragments into a single piece using Type IIs restriction enzymes and T4/T7 DNA ligase. This assembly is performed in vitro. Most commonly used Type IIs enzymes include BsaI, BsmBI and BbsI. Unlike standard Type II restriction enzymes like EcoRI and BamHI, these enzymes cut DNA outside of their recognition sites and therefore can create non-palindromic overhangs. Since 256 potential overhang sequences are possible, multiple fragments of DNA can be assembled by using combinations of overhang sequences. Golden Gate Assembly Schematic:
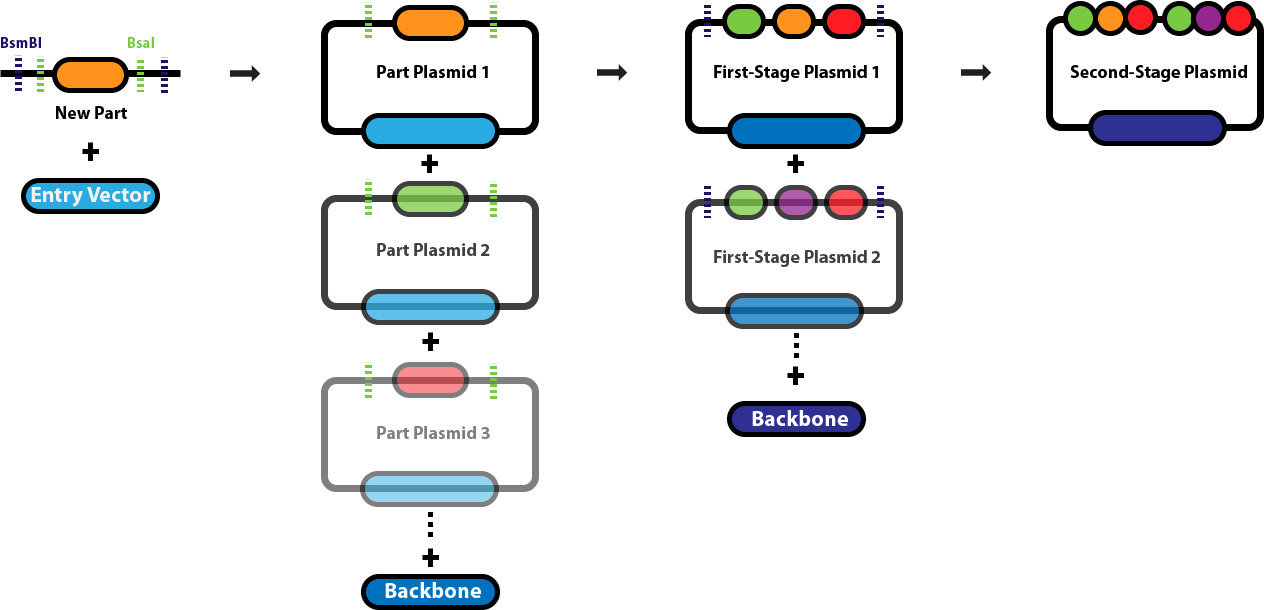
Golden Gate Assembly Links
Golden Gate Assembly - Start to Finish- Design a new part Design overhangs for the amplification of a new custom part to be assembled into a part plasmid.
- Assemble a new Part Plasmid Assemble a new custom part plasmid following amplification with part specific overhangs.
- Build a First-stage Plasmid (Transcriptional Unit) If you have all the requisite parts, you can build a complete plasmid with a single BsaI reaction.
- Build a Second-stage Plasmid Assemble multiple first-stage transcriptional unit plasmids.
- Tips, Tricks, and Troubleshooting Running into problems? Check out these potential solutions!
- Yeast Toolkit (YTK) - Derived from Lee, Dueber 2015 (old Golden Gate Assembly page!)
- Broad Host range Toolkit (BTK) - Derived from Leonard, Barrick 2018
Supplies
- 10× T4 DNA ligase buffer (NEB: M0202T)
- Why T4 DNA ligase buffer with T7 DNA ligase? T7 buffer contains PEG, which inhibits electroporation.
- T7 DNA ligase (NEB: M0318S)
- Why T7 DNA ligase instead of T4 DNA ligase? T7 does not ligate blunt ended DNA, yielding less off-target ligation.
- Restriction endonuclease BsaI (NEB: R0535) or BsmBI (NEB: R0580S)
- Competent cells
- SOC and liquid media
- Selective plates
Protocol
Use the Calculator Spreadsheet (linked at the bottom of the page) for planning your reactions!- Mix 20 fmol (1 nM final concentration) backbone and 40 fmol each insert of your DNA segments together. The volume of this mixture must be 16 µL.
- Add water to a final volume of 16 µl
- Add 2 µL of 10× T4 DNA ligase buffer. Mix by vortexing.
- Add 1 µL of BsaI or BsmBI and 1 µL of T7 DNA ligase. Mix by gently pipetting.
- Incubate the reaction for 30 temperature cycles (42°C for 5 min and then 16°C for 5 min), followed by a final 10 min incubation at 55°C. Alternate cycling protocols are described below, and may show better assemblies in some situations.
- Use 2 µL of this assembly reaction for electroporation or 4 µL for transforming chemically competent cells <- recommend change to 1uL.
References
- Lee ME, DeLoache WC, Cervantes B, Dueber JE. (2015) A Highly Characterized Yeast Toolkit for Modular, Multipart Assembly. ACS Synth. Biol. 4:975–986. Link
- Engler C, Kandzia R, Marillonnet S. (2008) A one pot, one step, precision cloning method with high throughput capability. PLoS ONE 3:e3647. Link
- "Golden Gate Assembly". New England Biolabs. Retrieved 8 June 2015. Link
Contributors
- Peng Geng
- Sean Leonard
- Kate Elston
- Dennis Mishler
- Jeffrey Barrick
-- Main.KateElston - 29 Jan 2018
| I | Attachment | History | Action | Size | Date | Who | Comment |
|---|---|---|---|---|---|---|---|
| |
BroadHostRange_Reaction_Calculator.xlsx | r1 | manage | 14.2 K | 2018-01-29 - 21:29 | KateElston | Golden Gate Reaction Calculator |
| |
golden_gate_diagram.ai | r1 | manage | 389.9 K | 2018-01-30 - 17:16 | KateElston |
Barrick Lab > ProtocolList > GoldenGateAssemblyProtocolsMainPage
Contributors to this topic  KateElston, DennisMishler, SeanLeonard, JuliePerreau, JeffreyBarrick
KateElston, DennisMishler, SeanLeonard, JuliePerreau, JeffreyBarrick
Topic revision: r4 - 2018-01-30 - 17:44:50 - Main.KateElston



 Mol Biosciences
Mol Biosciences The LTEE
The LTEE iGEM team
iGEM team NGS course
NGS course