Back to Golden Gate Protocols
Primer 1 (forward)
CAUTION In Primer 2, the priming site (dddd...) must be the reverse-complement of your part and you must use the suffix-R sequence for the Golden Gate part overlap because this is on the other strand. The overlap with the template can vary from the 20 base pairs that are shown according to the normal rules for designing good PCR primers. If calculating melting temperatures, be sure to only include the overlap region in your calculations, not the stuff that is being added to the ends!
Designing a new part
Golden Gate Assembly Step 1: Creating dsDNA encoding the part for cloning Two variations on preparing a dsDNA fragment with the proper restriction sites and Golden Gate overhangs are provided in the next sections. The end result is the same: a piece of DNA with the proper flanking regions for BsmBI cloning into the entry vector, while maintaining the BsaI sites used in first stage assembly – with proper overhangs for the type of part that you are designing.Golden Gate Part Design Reference:

Method 1: Amplifying a sequence with primers that add the required overhang regions
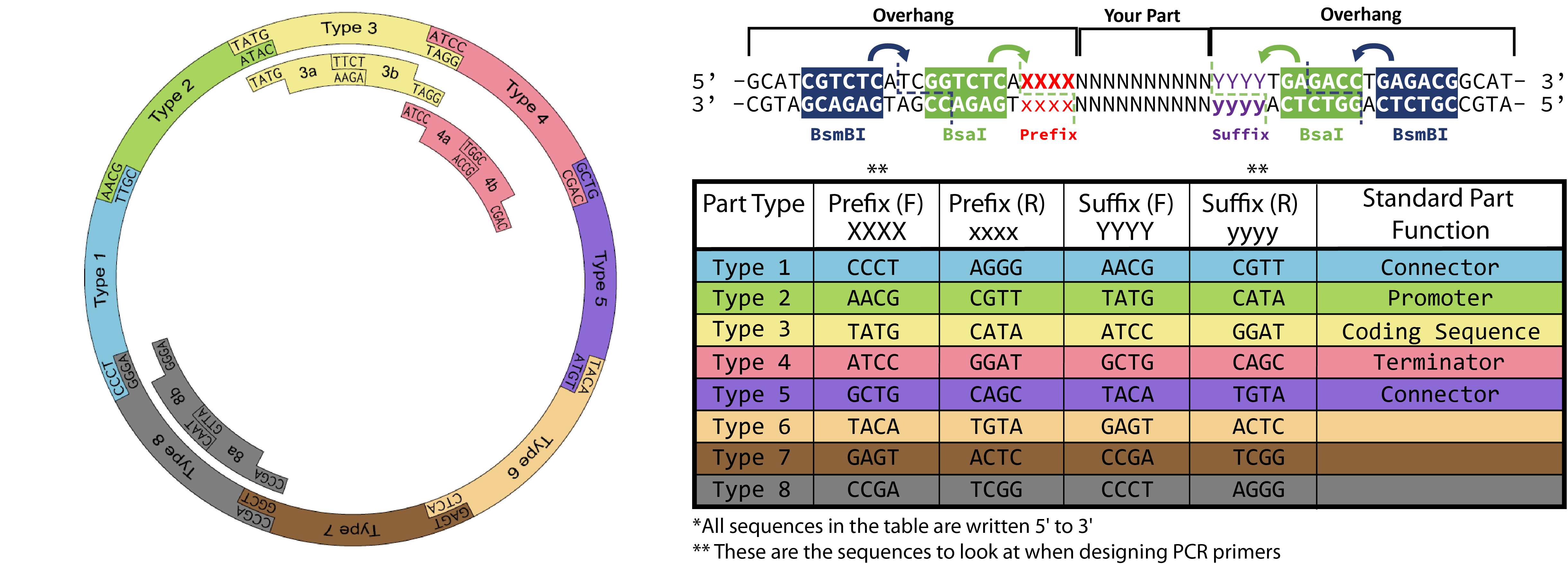
You need to order two primers that will anneal to your desired part sequence and contain overhang sequences necessary for proper Golden Gate Assembly. For easy primer design you can use the shiny app here: https://spleonard1.shinyapps.io/paRting/ Otherwise reference the table in the diagram at the top of the page to add the necessary prefix (XXXX)/suffix (yyyy) to the primer templates below! Type 3 overhang prefixes and type 2 suffixes (TATG) include start codons, so be careful to not add redundant start codons when designing primers for your type 3 part.Primer 1 (forward)
5'-GCATCGTCTCATCGGTCTCAXXXXUUUUUUUUUUUUUUUUUUUU-3'
Primer 2 (reverse)
5'-ATGCCGTCTCAGGTCTCAyyyyddddddddddddddddddd-3'
"UUUUU" = Upstream annealing region "ddddd" = Downsream annealing region (reverse complement)
CAUTION In Primer 2, the priming site (dddd...) must be the reverse-complement of your part and you must use the suffix-R sequence for the Golden Gate part overlap because this is on the other strand. The overlap with the template can vary from the 20 base pairs that are shown according to the normal rules for designing good PCR primers. If calculating melting temperatures, be sure to only include the overlap region in your calculations, not the stuff that is being added to the ends!
Example PCR Reaction
This protocol is for use with standard Phusion (or other high fidelity polymerase). Notice that there is an adjustment in the amount of dNTPs in the reaction compared to the standard protocol.
PCR (50ul reaction)
Set elongation time according to size of insert. Purify PCR products with a clean and concentrate kit if there is one band, and gel extract if there are more.
CAUTION The plasmid you are constructing in the entry vector (pYTK001) is camR, so if your template plasmid also encodes chloramphenicol resistance, you should DpnI digest after your PCR reaction, and probably also gel purify the fragment, as any residual plasmid may be transformed and lead to false-positive colonies.
- 10 µl 5x Buffer
- 3 µl dNTPs
- 2.5 µl FORWARD primer
- 2.5 µl REVERSE primer
- x µl template DNA (genomic = 50-250 ng, plasmid = 1 pg-10 ng )
- nfH2O to 49.5 µl
- then add 0.5 µl Phusion
Method 2: Synthesizing dsDNA containing the required overhang regions
When ordering a double-stranded piece of DNA to be synthesized, you can just append the sites needed for cloning in your order. Use this protocol when ordering a gBlock from IDT, for example.5'-GCATCGTCTCATCGGTCTCAXXXX YOUR PART YYYYTGAGACCTGAGACGGCAT-3'
3'-CGTAGCAGAGTAGCCAGAGTxxxx your part yyyyACTCTGGACTCTGCCGTA-5'
Replace XXXX with the prefix-F sequence for the part type you are designing ( see the table and notes above). Similarly, replace YYYY with the suffix-F sequence for the part.
In general, the amount of DNA synthesized is sufficient for cloning into the entry vector without further PCR amplification of a gBlock.
Back to Golden Gate Protocols
Barrick Lab > BroadHostRangeToolkit > ProtocolsBTKDesignANewPart
Topic revision: r15 - 2024-09-05 - 22:43:34 - Main.TylerDeJong