Barrick Lab :: Publications
Barrick Lab researchers *Equal contributions ^Corresponding author(s) Undergraduate researchers
| 2022 | |||
|---|---|---|---|
| 97. | Gómez-Garzón, C, Barrick, J.E., ^Payne, S.M. Disentangling the evolutionary history of Feo, the major ferrous iron transport system in bacteria. mBio 13:e03512-21. DOI: 10.1128/mbio.03512-21 PMID: 35012344 | ||
| 96. | Elston, K.M., Leonard, S.P., Geng, P., Bialik, S.B., Robinson, E., ^Barrick, J.E. (2022) Engineering insects from the endosymbiont out. Trends Microbiol. 30:79-96. DOI: 10.1016/j.tim.2021.05.004 PMID: 34103228 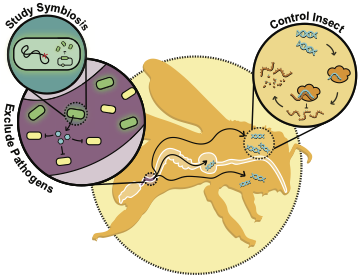 |
||
| 2021 | |||
| 95. | Deatherage, D.E., ^Barrick, J.E. (2021) High-throughput characterization of mutations in genes that drive clonal evolution using multiplex adaptome capture sequencing. _Cell Syst._*12*:79-96. DOI: 10.1016/j.cels.2021.08.011 PMID: 34536379 bioRxiv | ||
| 94. | Roots, C.T., Lukasiewicz, A., ^Barrick, J.E. (2021) OSTIR: open source translation initiation rate prediction. J. Open Source Softw. 6:3362. DOI: 10.21105/joss.03362 | ||
| 93. | Borin, J.M., Avrani, S., Barrick, J.E., Petrie, K.L., ^Meyer, J.R. Coevolutionary phage training leads to greater bacterial suppression and delays the evolution of phage resistance. Proc. Natl. Acad. Sci. U.S.A. 118:e2104592118. DOI: 10.1073/pnas.2104592118 PMID: 34083444 | ||
| 92. | McGuffie, M.C., ^Barrick, J.E. (2021) pLannotate: engineered plasmid annotation. Nucleic Acids Res. 49:W516-W522. DOI: 10.1093/nar/gkab374 PMID: 34019636
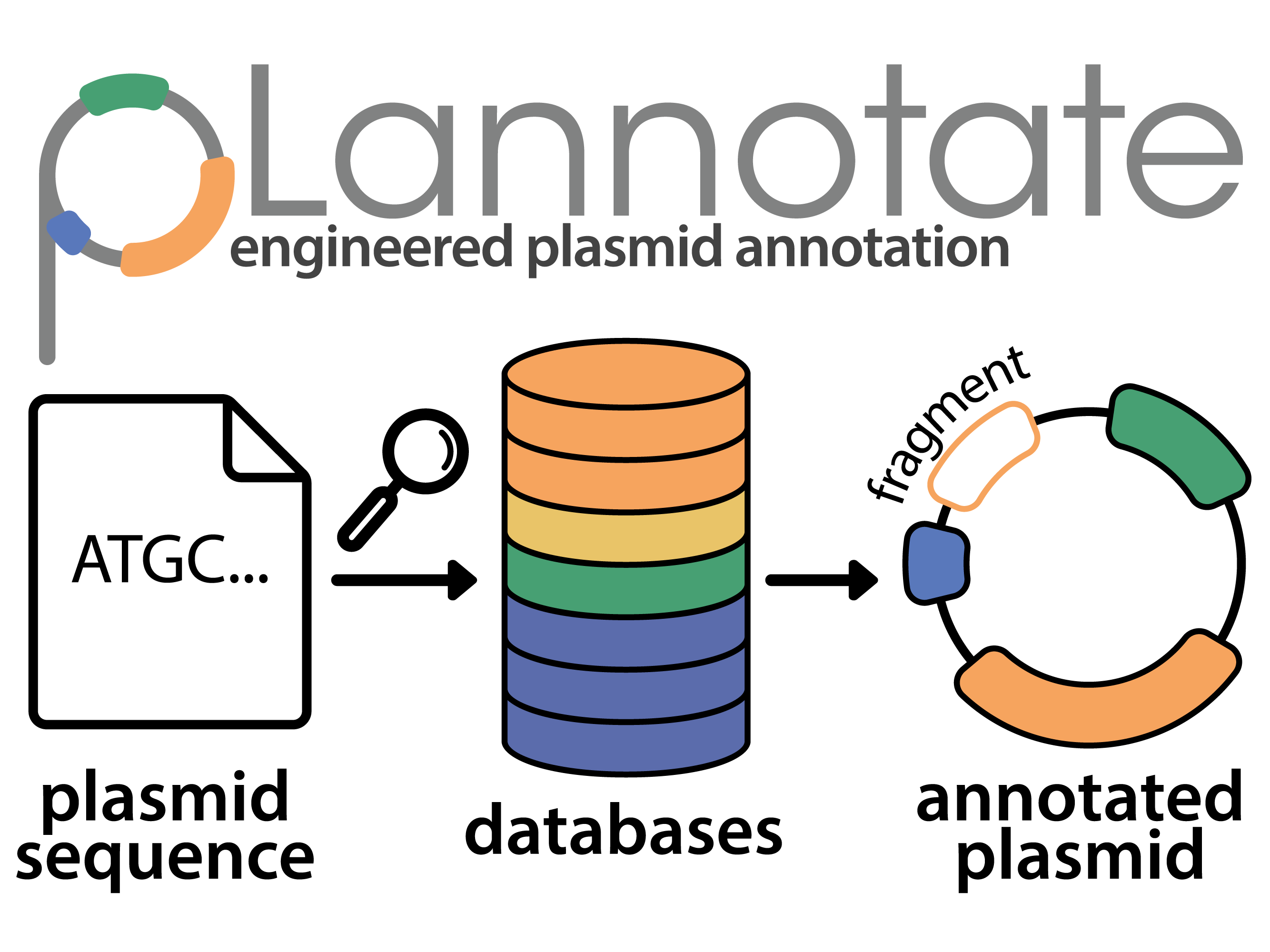 |
||
| 91. | ^Perreau, J., Patel, D.J., Anderson, H., Maeda, G.P, Elston, K.M., Barrick, J.E., Moran, N.A. Vertical transmission at the pathogen-symbiont interface: Serratia symbiotica and aphids. mBio 12:e00359-21. DOI: 10.1128/mBio.00359-21 PMID: 33879583 | ||
| 90. | Consuegra, J., Gaffé, J., Lenski, R. E., Hindré, T., Barrick, J. E., Tenaillon, O., Schneider, D. (2021) Insertion-sequence-mediated mutations both promote and constrain evolvability during a long-term experiment with bacteria. Nat. Commun. 12:980. DOI: 10.1038/s41467-021-21210-7 PMID: 33579917
|
||
| 89. | *Bazurto, J.V., *Riazi, S., D’Alton, S., Deatherage, D.E., Bruger, E.L., Barrick, J.E., ^Marx, C.J. (2021) Global transcriptional response of Methylorubrum extorquens to formaldehyde stress expands the role of EfgA and is distinct from antibiotic translational inhibition. Microorganisms 9:347. DOI: 10.3390/microorganisms9020347 PMID: 33578755 | ||
| 88. | ^Card, K.J., Thomas, M.D., Graves, J.L., Barrick, J.E., Lenski, R.E. (2021) Genomic evolution of antibiotic resistance is contingent on genetic background following a long-term experiment with Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 118: e2016886118. DOI: 10.1073/pnas.2016886118 PMID: 33441451 | ||
| 87. | *Gifford, I., *Dasgupta, A., ^Barrick, J.E. (2021) Rates of gene conversions between Escherichia coli ribosomal operons. G3 11:jkaa002. DOI: 10.1093/g3journal/jkaa002 PMID: 33585862 | ||
| 86. | Elston, K.M., Perreau, J., Maeda, G.P., Moran, N.A., ^Barrick, J.E. (2021) Engineering a culturable Serratia symbiotica strain for aphid paratransgenesis. Appl. Env. Microbiol. 87:e02245-20. DOI: 10.1128/AEM.02245-20 PMID: 33277267 |
||
| 2020 | |||
| 85. | Sher, A.A., *Jerome, J.P., Bell, J.A., Yu, J., Kim, H.Y., *Barrick, J.E., ^Mansfield, L.S. (2020) Experimental evolution of Campylobacter jejuni leads to loss of motility, rpoN (σ54) deletion and genome reduction. Front. Microbiol. 11:579989 DOI: 10.3389/fmicb.2020.579989 PMID: 33240235 | ||
| 84. | ^Barrick, J.E. (2020) Limits to predicting evolution: Insights from a long-term experiment with Escherichia coli. In: W. Banzhaf, B.H.C. Cheng, K. Deb, K.E. Holekamp, R.E. Lenski, C. Ofria, R.T. Pennock, W.F. Punch, D.J. Whittaker (eds.). Evolution in Action: Past, Present and Future: A Festschrift in Honor of Erik D. Goodman. pp 63-76. Springer Series on Genetic and Evolutionary Computation. Cham, Switzerland: Springer. Full Text DOI: 10.1007/978-3-030-39831-6_7 | ||
| 83. | ^Barrick, J.E., Deatherage D.E., ^Lenski, R.E. (2020) A test of the repeatability of measurements of relative fitness in the long-term evolution experiment with Escherichia coli. In: W. Banzhaf, B.H.C. Cheng, K. Deb, K.E. Holekamp, R.E. Lenski, C. Ofria, R.T. Pennock, W.F. Punch, D.J. Whittaker (eds.). Evolution in Action: Past, Present and Future: A Festschrift in Honor of Erik D. Goodman. pp 77-89. Springer Series on Genetic and Evolutionary Computation. Cham, Switzerland: Springer. Full Text DOI: 10.1007/978-3-030-39831-6_8 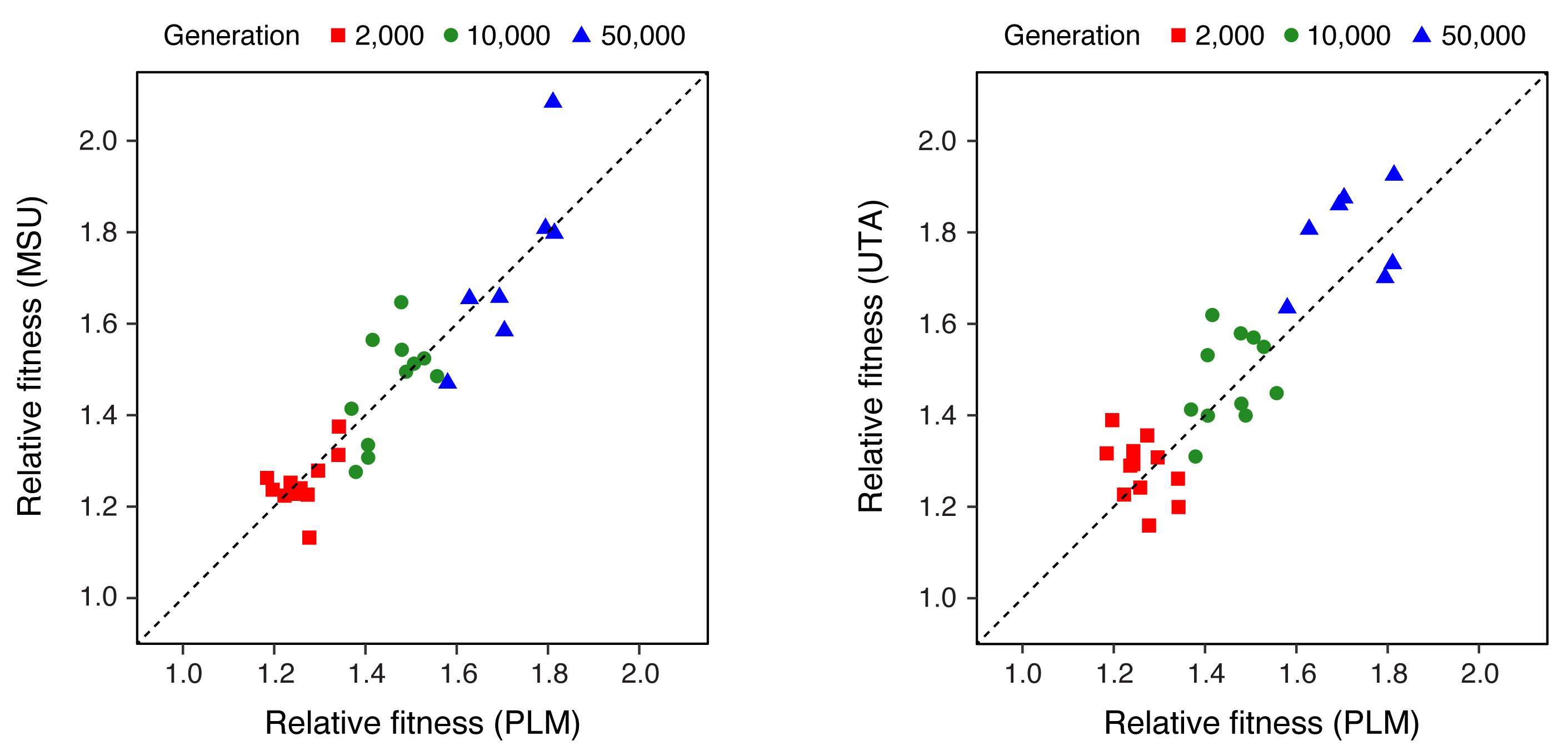 |
||
| 82. | ^*Blount, Z.D., ^*Maddamsetti, R., *Grant, N.A., Ahmed, S.T., Jagdish, T., Baxter, J.A._, Sommerfeld, B.A., Tillman, A., Moore, J., Slonczewski, J.L., Barrick, J.E., Lenski, R.E. (2020) Genomic and phenotypic evolution of Escherichia coli in a novel citrate-only resource environment. eLife 9:e55414. Full Text PMID: 32469311 | ||
| 81. | Suárez, G.A., Dugan, K.R., Renda, B.A., Leonard, S.P., Gangavarapu, L.S., ^Barrick, J.E. (2020) Rapid and assured genetic engineering methods applied to Acinetobacter baylyi ADP1 genome streamlining. Nucleic Acids Res. 48:4585–4600. Full Text PMID: 32232367 | ||
| 80. | Leonard, S.P., Powell, J.E., Perutka, J., Geng, P., Heckmann, L.C., Horak, R.D., Davies, B.W., Ellington, A.D., ^Barrick, J.E., and ^Moran, N.A. (2020) Engineered symbionts activate honey bee immunity and limit pathogens. Science 367:573–576. DOI, PMID: 32001655  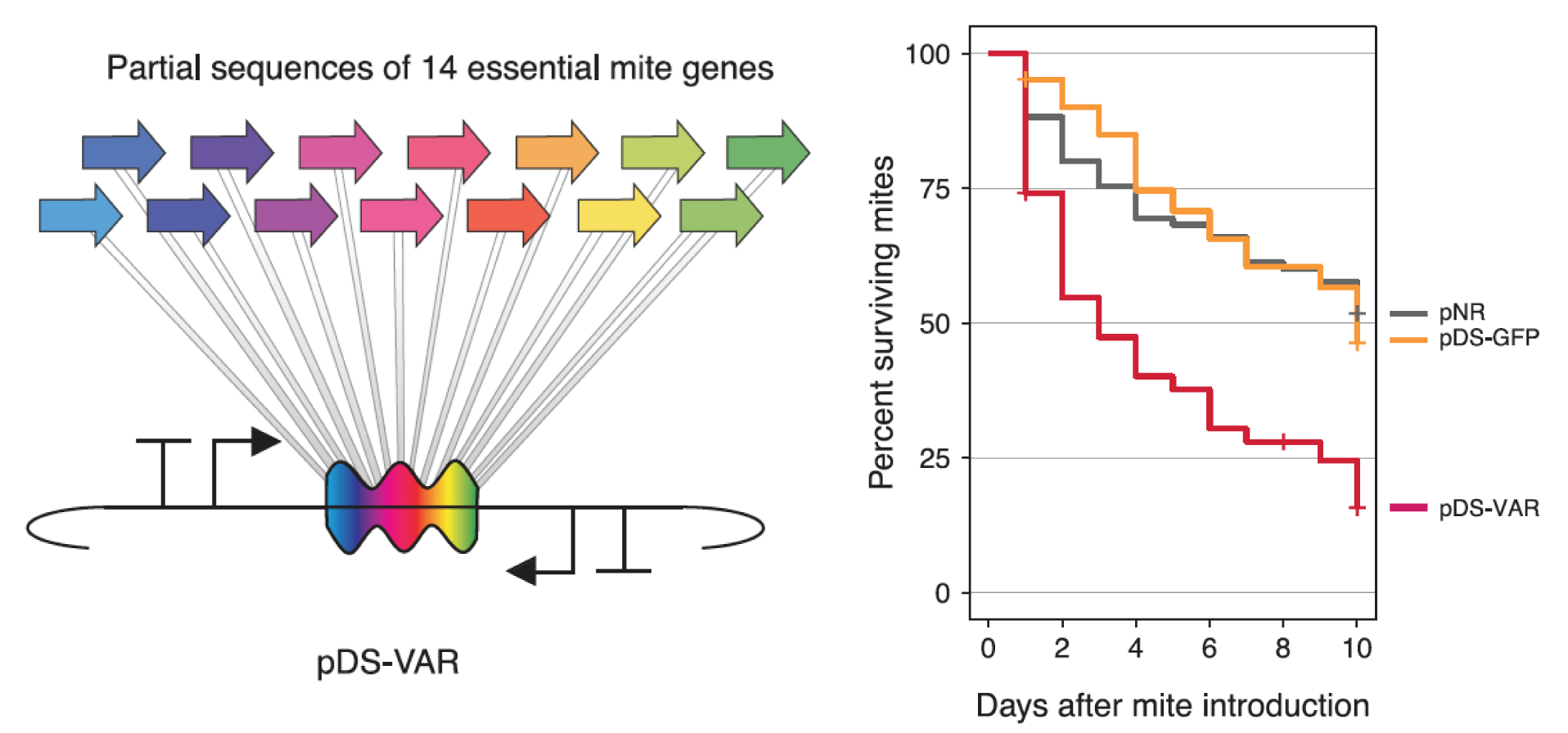 |
||
| 2019 | |||
| 79. | Zhang, X., Deatherage, D.E., Zheng, H., Georgoulis, S.J., ^Barrick, J.E. (2019) Evolution of satellite plasmids can stabilize the maintenance of newly acquired accessory genes in bacteria. Nat. Comm. 10:5809. Full Text, PMID: 31863068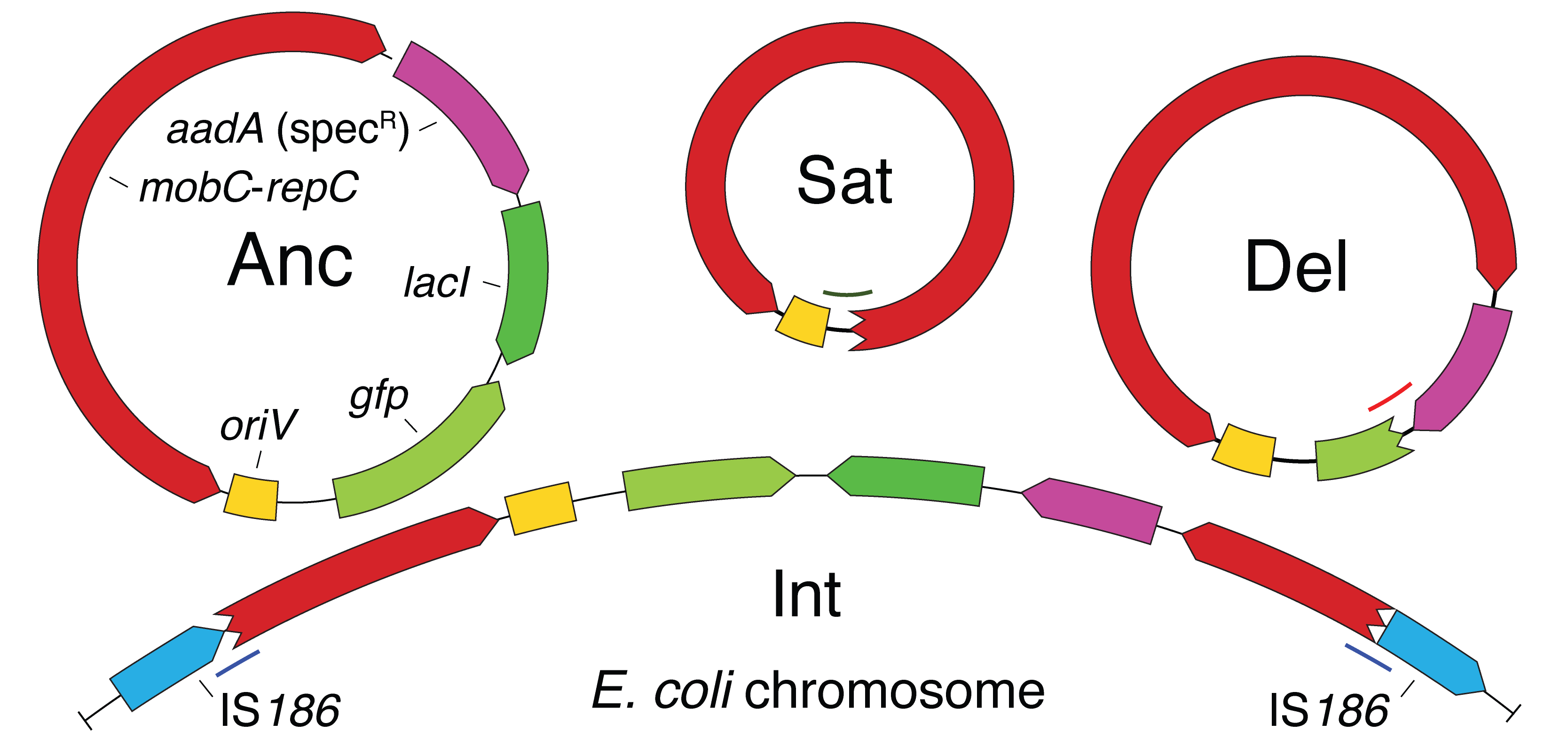 |
||
| 78. | Gutierrez, A.E., Shah, P., Rex, A.E., Nguyen, T.C., Kenkare, S.P., ^Barrick, J.E., ^Mishler, D.M. (2019) Bioassay for determining the concentrations of caffeine and individual methylxanthines in complex samples. Appl. Env. Microbiol. 85:e01965-19. PMID: 31540989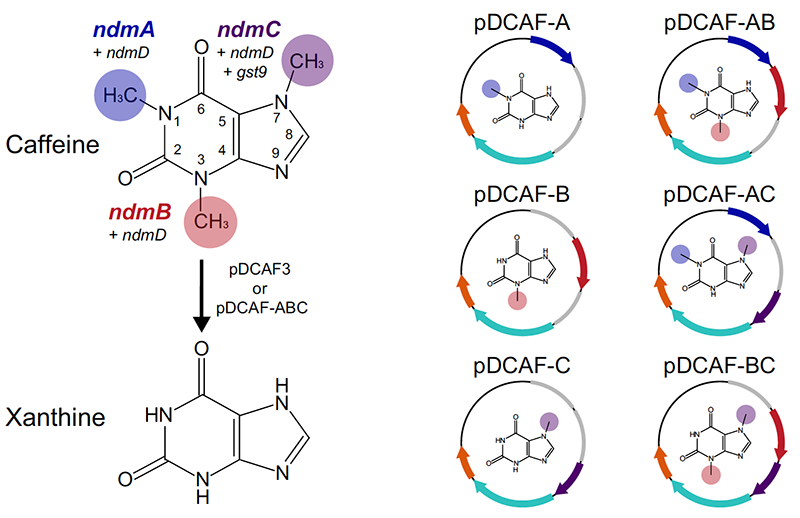 |
||
| 77. | Geng, P., Leonard, S.P., Mishler, D.M., ^Barrick, J.E. (2019) Synthetic genome defenses against selfish DNA elements stabilize engineered bacteria against evolutionary failure. ACS Synth. Biol. 8:521-531. Full Text, Preprint, PMID: 30703321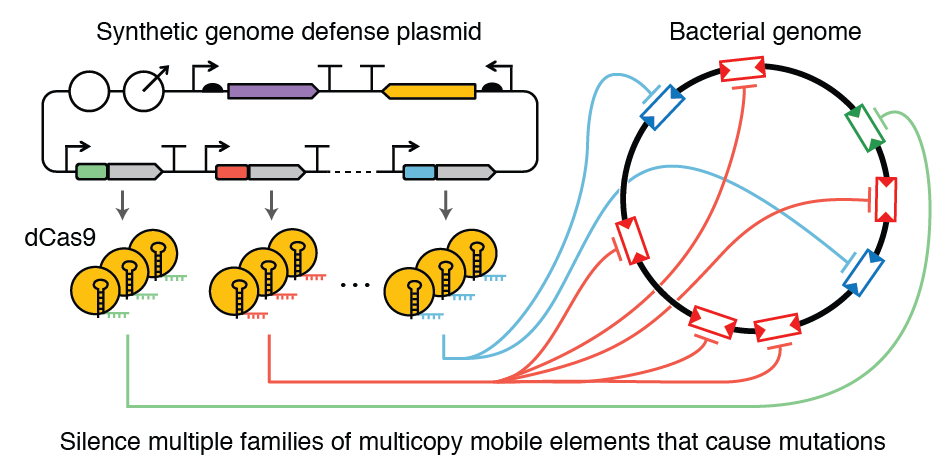 |
||
| 2018 | |||
| 76. | Deatherage, D.E., Leon, D., Rodriguez, Á.E., Omar, S., ^Barrick, J.E. (2018) Directed evolution of Escherichia coli with lower-than-natural plasmid mutation rates. Nucleic Acids Res. 46:9236-9250. Full Text, PMID: 30137492 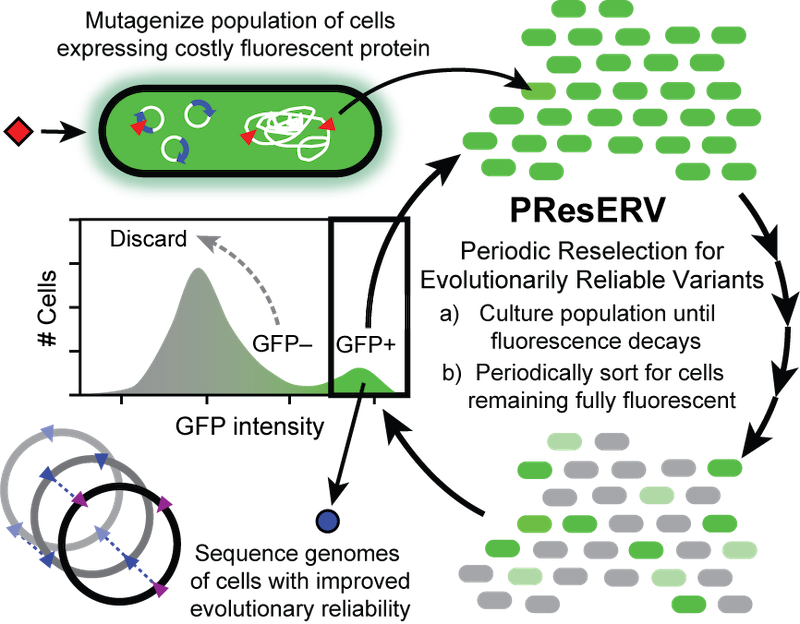 |
||
| 75. | *McGuffey, J.C., *Leon, D., Dhanji, E.Z., Mishler, D.M., ^Barrick, J.E. (2018) Bacterial production of gellan gum as a do-it-yourself alternative to agar. J. Microbiol. Biol. Educ. 19:182-184. Full Text, PMID: 29983852 |
||
| 74. | Leonard, S.P., Perutka, J., Powell, J.E., Geng, P., Richhart, D., Byrom, M., Kar, S., Davies, B.W., Ellington, A.D., ^Moran, N.A., ^Barrick, J.E. (2018) Genetic engineering of bee gut microbiome bacteria with a toolkit for modular assembly of broad-host-range plasmids. ACS Synth. Biol. 7:1279-1290. PMID: 29608282 |
||
| 73. | Leon, D., D’Alton, S., Quandt, E.M., ^Barrick, J.E. (2018) Innovation in an E. coli evolution experiment is contingent on maintaining adaptive potential until competition subsides. PLoS Genet. 14:e1007348. PMID: 29649242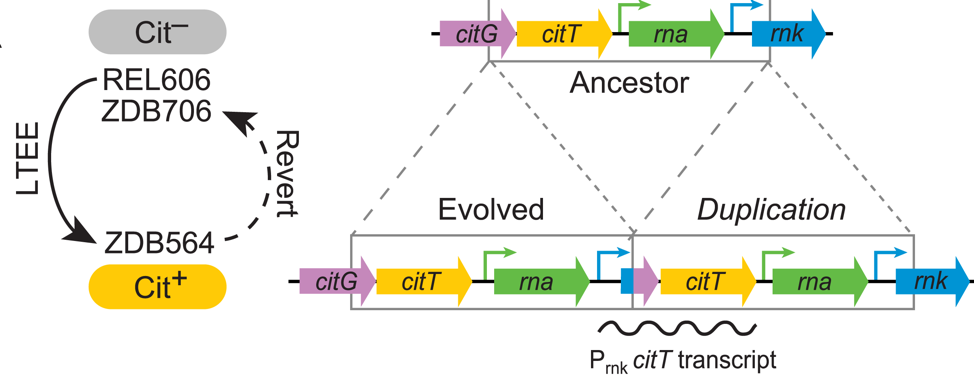 |
||
| 2017 | |||
| 72. | Good, B.H.*, McDonald, M.J.*, Barrick, J.E., Lenski R.E., Desai, M.M. (2017) The dynamics of molecular evolution over 60,000 generations. Nature 551:45-50. PMID: 29045390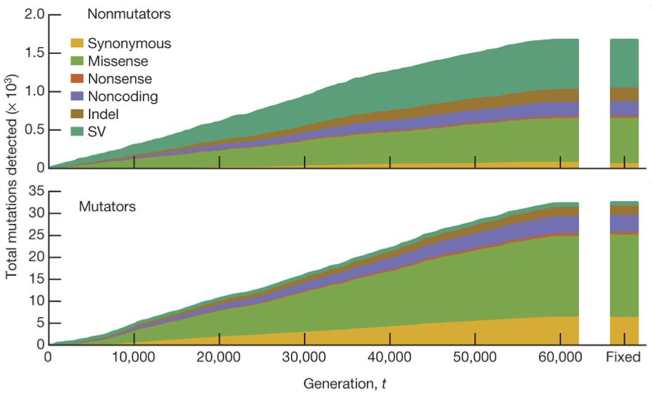 |
||
| 71. | Bull, J.J.*, Barrick, J.E.^* (2017) Arresting evolution. Trends Genet. 33:910-920. PMID: 29029851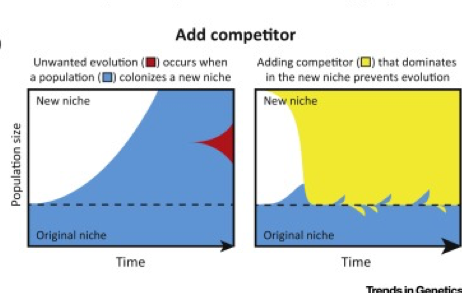 |
||
| 70. | ^Barrick, J.E. (2017) In future (cell) generations. “Voices” piece in What is the role of circuit design in the advancement of synthetic biology? Part 2. Cell Syst. 4:467-477. PMID: 28544877 | ||
| 69. | Suarez, G.A., Renda, B.A., Dasgupta, A., Barrick, J.E.^ (2017) Reduced mutation rate and increased transformability of transposon-free Acinetobacter baylyi ADP1-ISx. Appl. Env. Microbiol. 83: e01025-17. PMID: 28667117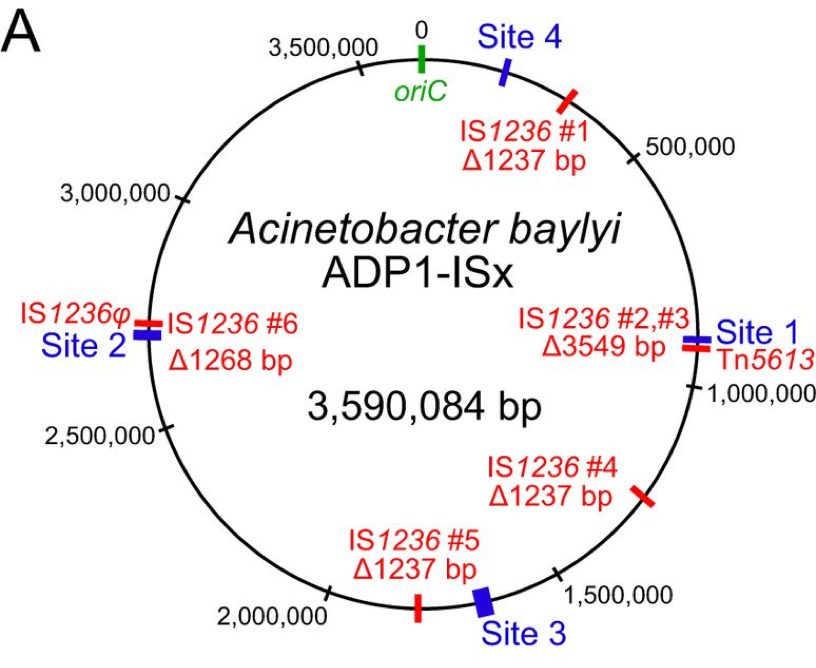 |
||
| 68. | Brown, C.W., Sridhara, V., Boutz, D.R., Person, M.D., Marcotte, E.M. Barrick, J.E., Wilke, C.O.^ (2017) Large-scale analysis of post-translational modifications in E. coli under glucose-limiting conditions. BMC Genomics 18:301. PMID: 28412930 | ||
| 67. | Caglar, M.U., Houser, J.R., Barnhart, C.S., Boutz, D.R., Carroll, S.M., Dasgupta, A., Lenoir, W.F., Smith, B.L., Sridhara, V., Sydykova, D.K., Vander Wood, D., Marx, C.J., Marcotte, E.M.^, Barrick, J.E.^, Wilke, C.O.^ (2017) The E. coli molecular phenotype under different growth conditions. Sci. Rep. 7:45303. PMID: 28417974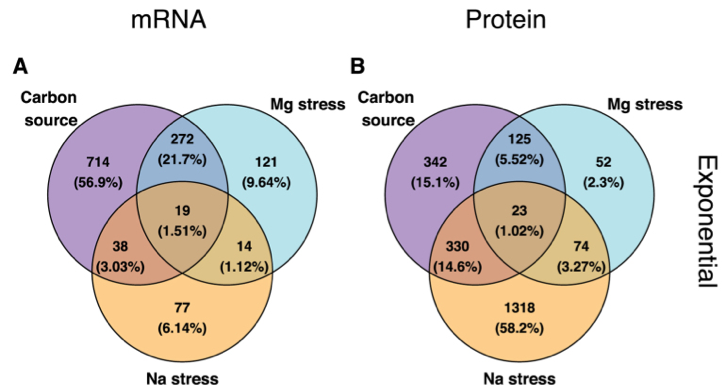 |
||
| 66. | Deatherage, D.E., Kepner, J.L., Bennett, A.F., Lenski, R.E.^, Barrick, J.E.^ (2017) Specificity of genome evolution in experimental populations of Escherichia coli evolved at different temperatures. Proc. Natl. Acad. Sci. U.S.A. 114:E1904-E1912. PMID: 28202733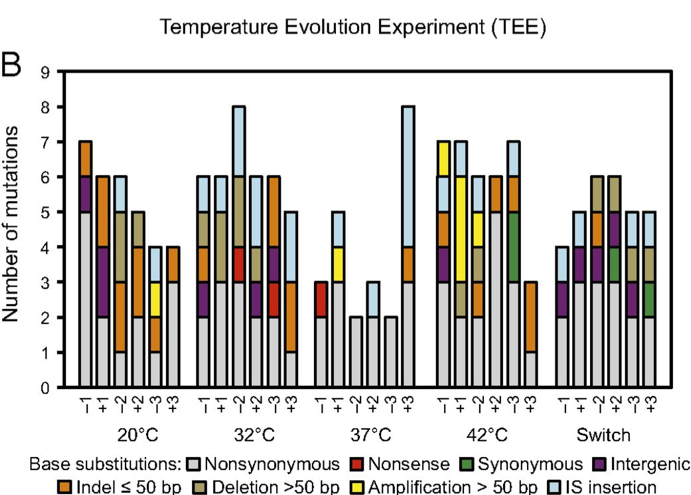 |
||
| 65. | Monk, J.W., Leonard, S.P., Brown, C.W., Hammerling, M.J., Mortensen, C., Gutierrez, A.E., Shin, N.Y., Watkins, E., Mishler, D.M.^, Barrick, J.E.^ (2017) Rapid and inexpensive evaluation of nonstandard amino acid incorporation in Escherichia coli. ACS Synth. Biol. 6:45-54. PMID: 27648665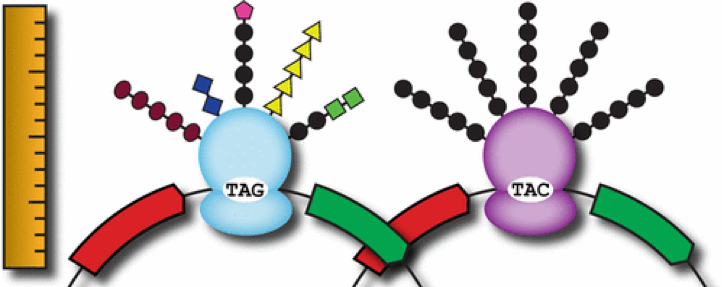 |
||
| 2016 | |||
| 64. | Renda, B.A., Chan, C., Parent, K.N., Barrick, J.E.^ (2016) Emergence of a competence reducing filamentous phage from the genome of Acinetobacter baylyi ADP1. J. Bacteriol. 198:3209-3219. PMID: 27645387 | ||
| 63. | Tenaillon, O.*, Barrick, J.E.*, Ribeck, N., Deatherage, D.E., Blanchard, J.L, Dasgupta, A., Wu, G.C., Wielgoss, S., Cruveiller, S., Medigue, C., Schneider, D., Lenski, R.E.^ (2016) Tempo and mode of genome evolution in a 50,000-generation experiment. Nature 536:165-170. PMID: 27479321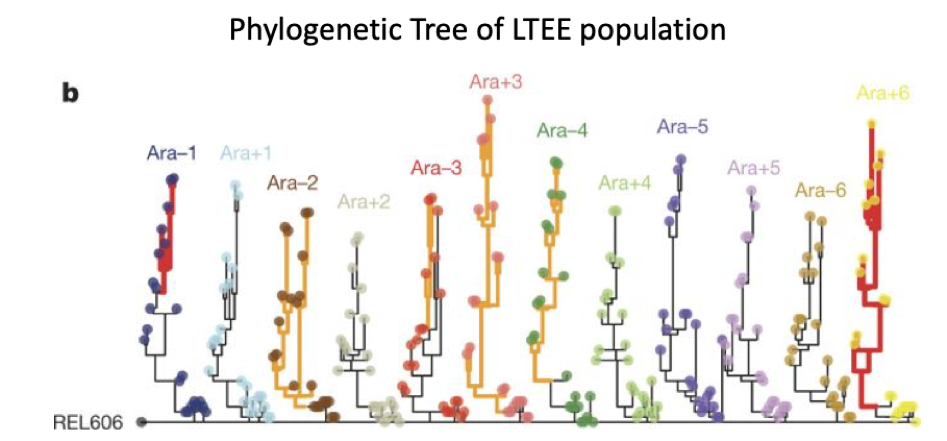 |
||
| 62. | Hammerling, M.J., Gollihar, J., Mortensen, C., Alnahhas, R.N., Ellington, A.D., Barrick, J.E. (2016) Expanded genetic codes create new mutational routes to rifampicin resistance in Escherichia coli. Mol. Biol. Evol. 33:2054-2063. PMID: 27189550 | ||
| 2015 | |||
| 61. | Quandt, E.M., Gollihar, J., Blount, Z.D., Ellington, A.D., Georgiou, G., Barrick, J.E. (2015) Fine-tuning citrate synthase flux potentiates and refines metabolic innovation in the Lenski evolution experiment. eLife 4:e09696. PMID: 26465114 |
||
| 60. | Houser, J.R., Barnhart, C., Boutz, D.R., Carroll, S.M., Dasgupta, A., Michener, J.K., Needham, B.D., Papoulas, O., Sridhara, V., Sydykova, D.K., Marx, C.J., Trent, S.M., Barrick, J.E.^, Marcotte, E.M.^, Wilke, C.O.^ (2015) Controlled measurement and comparative analysis of cellular components in E. coli reveals broad regulatory changes in response to glucose starvation. PLoS Comput. Biol. 11:e1004400. PMID:26275208 | ||
| 59. | Maddamsetti, R., Hatcher, P.J., Cruveiller, S., Médigue, C., Barrick, J.E., Lenski, R.E. (2015) Synonymous genetic variation in natural isolates of Escherichia coli does not predict where synonymous substitutions occur in a long-term experiment. Mol. Biol. Evol. 32:2897-2904. PMID:26199375 | ||
| 58. | Jack, B.R., Leonard, S.P., Mishler, D.M., Renda, B.A., Leon, D., Suárez, G.A., Barrick, J.E. (2015) Predicting the genetic stability of engineered DNA sequences with the EFM Calculator. ACS Synth. Biol. 4:939-943. PMID:26096262 | ||
| 57. | Perry, E.B.^, Barrick, J.E., Bohannan, B.J.M. (2015) The molecular and genetic basis of repeatable coevolution between Escherichia coli and bacteriophage T3 in a laboratory microcosm. PLoS ONE 10:e0130639. PMID:26114300. | ||
| 56. | Quandt, E.M., Summers, R.M., Subramanian, M.V., Barrick, J.E.^ (2015) Draft genome sequence of the bacterium Pseudomonas putida CBB5, which can utilize caffeine as a sole carbon and nitrogen source. Genome Announc. 3:e00640-15. PMID:26067973 | ||
| 55. | Maddamsetti, R., Lenski, R.E., Barrick, J.E. (2015) Adaptation, clonal interference, and frequency-dependent interactions in a long-term evolution experiment with Escherichia coli. Genetics 200:619-631. PMID:25911659 |
||
| 54. | Graves, J.L.^, Tajkarimi, M., Cunningham, Q., Campbell, A., Nonga, H., Harrison, S.H., Barrick, J.E. (2015) Rapid evolution of silver nanoparticle resistance in Escherichia coli. Front. Genet. 6:42. PMID:25741363 | ||
| 53. | Renda, B.A., Dasgupta, A., Leon, D., Barrick, J.E. (2015) Genome instability mediates the loss of key traits by Acinetobacter baylyi ADP1 during laboratory evolution. J. Bacteriol. 197:872-881. PMID:25512307 | ||
| 52. | Deatherage, D.E., Traverse, C.C., Wolf, L.N., Barrick, J.E. (2015) Detecting rare structural variation in evolving microbial populations from new sequence junctions using breseq. Front. Genet. 5:468. PMID:25653667 | ||
| 2014 | |||
| 51. | Sridhara, V., Meyer, A.G., Rai, P., Barrick, J.E., Ravikumar, P., Ségre, D., Wilke, C.O. (2014) Predicting growth conditions from internal metabolic fluxes in an in-silico model of E. coli. PLoS ONE 9:e114608. PMID:25502413 | ||
| 50. | Alnahhas, R.N.*, Slater, B.*, Huang, Y., Mortensen, C., Monk, J.W., Okasheh, Y., Howard, M.D., Gottel, N.R., Hammerling, M.J.^, Barrick, J.E.^ (2014) The case for decoupling assembly and submission standards to maintain a more flexible registry of biological parts. J. Biol. Eng. 8:28. PMID:25525459 | ||
| 49. | Barrick, J.E., Colburn, G., Deatherage D.E., Traverse, C.C., Strand, M.D., Borges, J.J., Knoester, D.B., Reba, A., Meyer, A.G. (2014) Identifying structural variation in haploid microbial genomes from short-read resequencing data using breseq. BMC Genomics 15:1039. PMID:25432719 | ||
| 48. | Raeside, C., Gaffé, J., Deatherage, D.E., Tenaillon, O., Briska, A.M., Ptashkin, R.N., Cruveiller, S., Médigue, C., Lenski, R.E., Barrick, J.E., Schneider, D. (2014) Large chromosomal rearrangements during a long-term evolution experiment with Escherichia coli. mBio 5:e01377-14. PubMed:25205090 | ||
| 47. | Deatherage, D.E., Barrick, J.E. (2014) Identification of mutations in laboratory-evolved microbes from next-generation sequencing data using breseq. Methods Mol. Biol. 1151:165-188. PubMed:24838886 | ||
| 46. | Renda, B.A.*, Hammerling, M.J.*, Barrick, J.E. (2014) Engineering reduced evolutionary potential for synthetic biology. Mol. Biosyst. 10:1668-1678. PubMed:24556867 | ||
| 45. | Hammerling, M.J., Ellefson, J.W., Boutz, D.R., Marcotte, E.M., Ellington, A.D., Barrick, J.E. (2014) Bacteriophages use an expanded genetic code on evolutionary paths to higher fitness. Nat. Chem. Biol. 10:178-180. PubMed:24487692 | ||
| 44. | Quandt, E.M., Deatherage, D.E., Ellington, A.D., Georgiou, G., Barrick, J.E. (2014) Recursive genomewide recombination and sequencing reveals a key refinement step in the evolution of a metabolic innovation in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 111:2217-2222. PubMed:24379390 |
||
| 2013 | |||
| 43. | Hammerling, M.J., Gottel, N.R., Alnahhas, R.N., Slater, B., Huang, Y., Okasheh, Y., Howard, M., Mortensen, C., Monk, J., Detelich, M., Lannan, R.S., Pitaktong, A., Weaver, E., Das, S., Barrick, J.E. (2013) BBF RFC95: Open Sequence Initiative: a part submission standard to complement modern DNA assembly techniques. «BioBricks Foundation» «DSpace» | ||
| 42. | Barrick, J.E., Lenski, R.E. (2013) Genome dynamics during experimental evolution. Nat. Rev. Genet. 14:827-834. «PubMed» | ||
| 41. | Summers, R.M., Seffernick, J.L., Quandt, E.M., Yu, C.L., Barrick, J.E., Subramanian, M.V. (2013) Caffeine Junkie: an unprecedented GST-dependent oxygenase required for caffeine degradation by P. putida CBB5. J. Bacteriol. 195:3933-3939. «PubMed» | ||
| 40. | Quandt, E.M., Hammerling, M.J., Summers, R.M., Otoupal, P.B., Slater, B., Alnahhas, R.N., Dasgupta, A., Bachman, J.L., Subramanian, M.V., Barrick, J.E. (2013) Decaffeination and measurement of caffeine content by addicted Escherichia coli with a refactored N-demethylation operon from Pseudomonas putida CBB5. ACS Synth. Biol. 2:301-307. «PubMed» | ||
| 39. | Han, P., Niestemski, L.R., Barrick, J.E., Deem, M.W. (2013) Physical model of the immune response of bacteria against bacteriophage through the adaptive CRISPR-Cas immune system. Phys. Biol. 10:025004. «PubMed» | ||
| 38. | Wielgoss, S.*, Barrick, J.E.*,Tenaillon, O.*, Wiser, M.J., Dittmar, W.J., Cruveiller, S., Chane-Woon-Ming, B., Médigue, C., Lenski, R.E., Schneider, D. (2013) Mutation rate dynamics in a bacterial population reflect tension between adaptation and genetic load. Proc. Natl. Acad. Sci. U.S.A. 110:222-227. «PubMed» | ||
| 2012 | |||
| 37. | Blount, Z.D., Barrick, J.E., Davidson, C.J., Lenski, R.E. (2012) Genomic analysis of a key innovation in an experimental E. coli population. Nature 489:513-518. «PubMed» | ||
| 36. | Jerome, J.P., Klahn, B., Bell, J.A., Barrick, J.E., Brown, C.T., Mansfield, L.S. (2012) Draft genome sequences of two Campylobacter jejuni clinical isolates, NW and D2600. J.Bact. 194:5707-5708. | ||
| 35. | Reba, A., Meyer, A.G., Barrick, J.E. (2012) Computational tests of a thermal cycling strategy to isolate more complex functional nucleic acid motifs from random sequence pools by in vitro selection. In: C. Adami et al. (eds.). Artificial Life XIII: Proceedings of the Thirteenth International Conference on the Synthesis and Simulation of Living Systems. pp 473-480. Cambridge, MA: MIT Press. Awarded Best Synthetic Biology Paper. «Full Text» | ||
| 34. | Kholmanov, I.N., Stoller, M.D., Edgeworth, J., Lee, W.H., Li, H., Lee, J., Barnhart, C., Potts, J.R., Piner, R., Akinwande, D., Barrick, J.E., Ruoff, R.S. (2012) Nanostructured hybrid transparent conductive films with antibacterial properties. ACS Nano 6:5157-5163. «PubMed» | ||
| 33. | Wolf, L.N., Barrick, J.E. (2012) Tracking winners and losers in E. coli evolution experiments. Microbe Magazine 2 (3):124-128. «Article» | ||
| 32. | Meyer, J.R., Dobias, D.T., Weitz, J.S., Barrick, J.E., Quick, R.T., Lenski, R.E. (2012) Repeatability and contingency in the evolution of a key innovation in phage lambda. Science 335:428-432. «PubMed» | ||
| 2011 | |||
| 31. | Nahku, R., Peebo, K., Valgepea, K., Barrick, J.E., Adamberg, K., Vilu, R. (2011) Stock culture heterogeneity rather than new mutational variation complicates short-term cell physiology studies of Escherichia coli K-12 MG1655 in continuous culture. Microbiology 157:2604-2610. «PubMed» | ||
| 30. | Wielgoss, S., Barrick, J.E., Tenaillon, O., Cruveiller, S., Chane-Woon-Ming, B., Médigue, C., Lenski, R.E., Schneider, D. (2011) Mutation rate inferred from synonymous substitutions in a long-term evolution experiment with Escherichia coli. G3: Genes, Genomes, Genetics 1:183-186. «PubMed» |
||
| 29. | Woods, R.J.*, Barrick, J.E.*^, Cooper, T.F., Shrestha, U., Kauth, M.R., Lenski, R.E.^ (2011) Second-order selection for evolvability in a large Escherichia coli population. Science 331:1433-1436. «PubMed» «Press Links» «Science Podcast» «Faculty of 1000» «Erratum» | ||
| 28. | Jerome, J.P., Bell, J.A., Plovanich-Jones, A.E., Barrick, J.E., Brown, C.T., Mansfield, L.S. (2011) Standing genetic variation in contingency loci drives the rapid adaptation of Campylobacter jejuni to a novel host. PLoS ONE 6:e16399. «PubMed» «Blog Article» | ||
| 2010 and earlier | |||
| 27. | Barrick, J.E., Kauth, M.R., Strelioff, C.C., and Lenski, R.E. (2010) Escherichia coli rpoB mutants have increased evolvability in proportion to their fitness defects. Mol. Biol. Evol. 27:1338-1347. «PubMed» | ||
| 26. | Barrick, J.E.*, Yu, D.S.*, Yoon, S.H., Jeong, H, Oh, T.K., Schneider, D., Lenski, R.E., and Kim, J.F. (2009) Genome evolution and adaptation in a long-term experiment with Escherichia coli. Nature. 461:1243-1247. «PubMed»«Faculty of 1000» |
||
| 25. | Barrick, J.E. and Lenski, R.E. (2009) Genome-wide mutational diversity in an evolving population of Escherichia coli. Cold Spring Harbor Symp. Quant. Biol. 74:119-129. «PubMed» | ||
| 24. | Barrick, J.E. (2009) Predicting riboswitch regulation on a genomic scale. In Riboswitches: Methods and Protocols. (ed. A. Serganov), pp. 1-13. Humana Press, New York. «PubMed» |
 |
|
| 23. | Regulski, E.E., Moy, R.H., Weinberg, Z., Barrick, J.E., Yao, Z., Ruzzo, W.L., and Breaker, R.R. (2008) A widespread riboswitch candidate that controls bacterial genes involved in molybdenum cofactor and tungsten cofactor metabolism. Mol. Microbiol. 68:918-932. «PubMed» | ||
| 22. | Weinberg, Z., Regulski, E.E., Hammond, M.C., Barrick, J.E., Yao, Z., Ruzzo, W.L., and Breaker, R.R. (2008) The aptamer core of SAM-IV riboswitches mimics the ligand-binding site of SAM-I riboswitches. RNA 14:822-828. «PubMed» | ||
| 21. | Barrick, J.E. and Breaker R.R. (2007) The distributions, mechanisms, and structures of metabolite-binding riboswitches. Genome Biology 8:R239. «PubMed» | ||
| 20. | Weinberg, Z., Barrick, J.E., Yao, Z., Roth, A., Kim, J.N., Gore, J., Wang, J.X., Lee, E.R., Block, K.F., Sudarsan, N, Neph, S., Tompa, M., Ruzzo, W.L., Breaker, R.R. (2007) Identification of 22 candidate structured RNAs in bacteria using the CMfinder comparative genomics pipeline. Nucleic Acids Res. 35:4809-4819 «PubMed» | ||
| 19. | Yao, Z., Barrick, J.E., Weinberg, Z., Neph, S., Breaker, R.R., Tompa, M., Ruzzo, W.L. (2007) A computational pipeline for high-throughput discovery of cis-regulatory noncoding RNA in prokaryotes. PLOS Comput. Biol. 3:e126. «PubMed» | ||
| 18. | Roth, A., Winkler, W.C., Regulski, E.E., Lee, B.W., Lim, J., Jona, I., Barrick, J.E., Ritwik, A., Kim, J.N., Welz, R., Iwata-Reuyl D., Breaker R.R. (2007) A riboswitch selective for the queuosine precursor preQ1 contains an unusually small aptamer domain. Nat. Struct. Mol. Biol. 14 (4):308-317. «PubMed» | ||
| 17. | Barrick, J.E. and Breaker R.R. (2007) The Power of Riboswitches. Scientific American 296 (1):50-57. «PubMed» | ||
| 16. | Lenski, R.E., Barrick, J.E., and Ofria, C. (2006) Balancing Robustness and Evolvability. PLoS Biology 4:e428. «PubMed» | ||
| 15. | Puerta-Fernandez, E., Barrick, J.E., Roth, A., and Breaker, R.R. (2006) Identification of a new, non-coding RNA in extremophilic eubacteria. Proc. Natl. Acad. Sci U.S.A 103:19490-19495. «PubMed» | ||
| 14. | Sudarsan N., Hammond M.C., Block K.F., Welz R., Barrick J.E., Roth A., Breaker R.R. (2006) Tandem riboswitch architectures exhibit complex gene control functions. Science 314:300-304. «PubMed» | ||
| 13. | Corbino, K.A., Barrick, J.E., Lim, J., Welz, R., Tucker, B.J., Puskarz, I., Mandal, M., Rudnick, N.D., and Breaker, R.R. (2005) Evidence for a second class of S-adenosylmethionine riboswitches and other regulatory RNA motifs in alpha-proteobacteria. Genome Biol. 6: R70. «PubMed» | ||
| 12. | Barrick, J.E., Sudarsan, N., Weinberg, Z., Ruzzo, W.L., and Breaker, R.R. (2005) 6S RNA is a widespread regulator of eubacterial RNA polymerase that resembles an open promoter. RNA 11:774-784. «PubMed» | ||
| 11. | Mandal, M., Lee, M., Barrick, J.E., Weinberg, Z., Emilsson, G.M., Ruzzo, W.L., and Breaker, R.R. (2004) A glycine-dependent riboswitch that uses cooperative binding to control gene expression. Science 306:275-279. «PubMed» | ||
| 10. | Barrick, J.E., Corbino, K.A., Winkler, W.C., Nahvi, A., Mandal, M., Collins, J., Lee, M., Roth, A., Sudarsan, N., Jona, I., Wickiser, J.K., and Breaker, R.R. (2004) New RNA motifs suggest an expanded scope for riboswitches in bacterial genetic control. Proc. Natl. Acad. Sci. U.S.A. 101:6421-6426. «PubMed» | ||
| 9. | Nahvi, A., Barrick, J.E., and Breaker, R.R. (2004) Coenzyme B12 riboswitches are widespread genetic control elements in prokaryotes. Nucleic Acids Res. 32:143-150. «PubMed» | ||
| 8. | Mandal, M., Boese, B., Barrick, J.E., Winkler, W.C., and Breaker, R.R. (2003) Riboswitches control fundamental biochemical pathways in Bacillus subtilis and other bacteria. Cell 113:577-586. «PubMed» | ||
| 7. | Sudarsan, N., Barrick, J.E., and Breaker, R.R. (2003) Metabolite-binding RNA domains are present in the genes of eukaryotes. RNA 9:644-647. «PubMed» | ||
| 6. | Winkler, W.C., Nahvi, A., Sudarsan, N., Barrick, J.E., and Breaker, R.R. (2003) An mRNA structure that controls gene expression by binding S-adenosylmethionine. Nat. Struct. Biol. 10:701-707. «PubMed» | ||
| 5. | Barrick, J.E., and Roberts, R.W. (2003) Achieving specificity in selected and wild-type N peptide–RNA complexes: The importance of discrimination against noncognate RNA targets. Biochemistry 42:12998-13007. «PubMed» | ||
| 4. | Barrick, J.E., and Roberts, R.W. (2002) Sequence analysis of an artificial family of RNA-binding peptides. Protein Sci. 11:2688-2696. «PubMed» | ||
| 3. | Barrick, J.E., Takahashi, T.T., Ren, J.S., Xia, T.B., and Roberts, R.W. (2001) Large libraries reveal diverse solutions to an RNA recognition problem. Proc. Natl. Acad. Sci. U.S.A. 98:12374-12378. «PubMed» | ||
| 2. | Barrick, J.E., Takahashi, T.T., Balakin, A., and Roberts, R.W. (2001) Selection of RNA-binding peptides using mRNA-peptide fusions. Methods 23:287-293. «PubMed» | ||
| 1. | Liu, R.H., Barrick, J.E., Szostak, J.W., and Roberts, R.W. (2000) Optimized synthesis of RNA-protein fusions for in vitro protein selection. Methods Enzymol. 318:268-293. «PubMed» | ||
Ph.D Thesis
| Barrick JE. (2006) Advisor: Ronald R. Breaker. Discovering and defining metabolite-binding riboswitches and other structured regulatory RNA motifs in bacteria. «PDF» |  |
This topic: Lab > WebHome > PublicationList
Topic revision: r233 - 2022-01-25 - SarahBialik