| |
| META TOPICPARENT |
name="QPCR" |
<<Return to qPCR page
Reference Gene qPCR
Goals
- Determine which of your reference genes you are going to normalize to.
Why am I doing this? |
|
<
< | Reference genes need to be stable across your control and experimental samples in order to be useful. If expression between the two differs, you will be normalizing to two different values and that is worthless. This is a common omission. Reference genes must be validated. |
>
> | Reference genes need to be stable across your control and experimental samples in order to be useful. If expression between the two differs, you will be normalizing to two different values, reference genes should always be validated before you use them in your experiments! |
| |
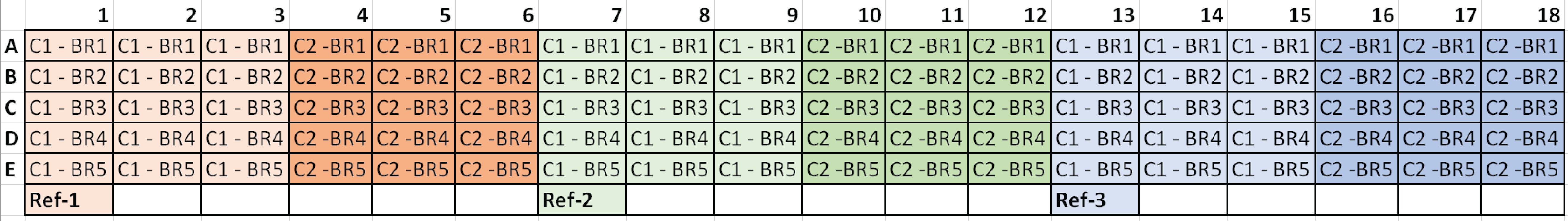
Typical plate setup for three candidate reference genes: |
|
<
< | |
>
> |  |
| | |
|
<
< | A |
>
> | *C1 = Condition 1, C2 = Condition 2, and BR# = Biological Replicate |
|
<
< | |
| | |
|
<
< | C1BR1 |
>
> | Template Material_ |
|
<
< | |
| | |
|
<
< | C1BR1 |
>
> | |
|
<
< | |
| | |
|
<
< | C1BR1 |
>
> | What you are looking for |
|
<
< | |
| | |
|
<
< | |
>
> |
- Technical replicates with a standard deviation below 0.2 (this confirms the accuracy of your results).
- At least 2 reference primer sets that show no significant difference between control and experimental conditions. The standard deviation of the Cqs for all biological replicates should be low. (Preferrably Less than 0.5)
|
|
>
> |
- If you are seeing differences between replicates or perhaps conditions, the main issue is likely from setting up the qPCR plate itself. The first few times you run qPCR the results can be messy and sometimes you just have to redo things to get accurate results. That being said, if you're a qPCR pro, or if you keep getting the same results over and over you may have to design primers for a new reference.
|
| | |
|
<
< | |
| | |
|
<
< | C1BR1 |
>
> | <<Return to qPCR page |
|
<
< | |
| | |
|
<
< | |
| | |
|
<
< | |
| | |
|
<
< | |
| | |
|
<
< | |
| | |
|
<
< | C1BR2 |
>
> |
| META FILEATTACHMENT |
attachment="Refprimerplate.png" attr="h" comment="" date="1582235087" name="Refprimerplate.png" path="Refprimerplate.png" size="102909" stream="Refprimerplate.png" tmpFilename="/usr/tmp/CGItemp43424" user="KateElston" version="2" |
|
|
<
< |
Conditions
What you are looking for
- Technical replicates with a standard deviation below 0.2 (this is arbitrary and most of your replicates will be below 0.1. If you do enough qPCR, you will eventually become obsessed with how low you can get this number).
- At least 2 reference primer sets that show no significant difference between control and experimental conditions. The standard deviation of the Cqs for all biological replicates should be low. (Preferrably Less than 0.5)
- If you are seeing differences between replicates or perhaps conditions, you may be asking the question “how do I know if there’s really a difference, it could be something else like loading or RT-PCR efficiency? I am not controlling for any of these by normalizing to anything!”. The answer is if you prepared good quality RNA and loaded exactly the same amount of RNA into a well-prepared reverse transcription, there should be very little (less than 1 Cq) difference between biological replicates of the same condition, assuming that condition itself is reproducible. If the biological variation is truly large between replicates, you’ll have to pick the best you can.
<<Return to qPCR page |
| | |

