
Difference: RNASeq (1 vs. 6)
Revision 62021-05-10 - GabrielSuarez
Revision 52018-01-23 - SimonDAlton
RNAseq Library PreparationUse RNA from RNASnap and Zymo coumn purification as outlined on wiki.rRNA DepletionUse the Ribozero (RZ) rRNA Removal Kit (Gram-negative bacteria) from Illumina. We assayed the effectiveness of the RZ kit: using inputs of up to 5ug, rRNA was reduced from >80% of total cell RNA to less than 1%. Protocol as follows: *Cool a benchtop centrifuge to 40C, place 100% ethanol and 70% ethanol on ice (store chilled at -200C). Set a waterbath to 500C* DO NOT PUT MAGNETIC BEADS ON ICE. All RZ components can be kept at room temperature during the procedure. Protocol: Batch wash magnetic beads:
ETOH precipitation purificationThe pellets - including the final pellet - may not be easily visible, so keep track of which 'corner' of the centrifuge tube is facing outwards in centrifuge. Protocol:
RNA FragmentationUse NEB Fragmentation Buffer. The RNA frag buffer conditions and high temperature cause RNA fragmentation. Fragment size is determined by time exposed to both. For tight control of fragmentation, add the buffer to the RNA in a strip tube, seal, resuspend tube by thoroughly flicking strip, centrifuge briefly and place in a pre-heated PCR cycler. Likewise, at the end of the run immediately cool on ice, collect any condensation by centrifugation and then add Stop Buffer. For E.coli, in our hands, 2 minutes at 94 degrees produces a final PCR product smear (see final steps) of between 150bp and 500bp (>98% of product between 150bp and 300bp). This should be determined experimentally from lab to lab. Protocol:
Kinase Treatment of Fragmented mRNAA master mix of buffer, T4 PNK and ATP can be made and 6ul added to n empty PCR strip tube wells (on ice) during the above ethanol precipitation's final 5 minute drying phase (see note above). Then resuspended RNA can be added to the reaction mix and thoroughly mixed immediately. Protocol:
Library Preparation
Use the NEBNext Small RNA Library Prep Set for Illumina (Multiplex Compatible), catalogue #E7330S/L (24/96 rxns).
The entire protocol is carried out in a single tube per sample, each subsequent reaction mixture being added to the previous product. Care should therefore be taken after each step to cool the products on ice and collect them by brief centrifugation so as to maintain reaction volume.
Note that many of the below steps require pre-heated blocks or transfers between temperatures. Plan accordingly.
Due to the low concentration of RNA following RZ, we routinely dilute the adapters, primers etc that come in the kit 1 in 2 (e.g. 1ul H2O + 1ul primer)
Protocol:
3’ SR Adaptor Ligation | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3' ligation buffer | 10ul |
| 3' ligation enzyme mix | 3ul |
| Total cumulative reaction volume | 20ul |
- Seal, flick thoroughly to mix, collect by centrifugation.
- Incubate for 1 hour at 250C (do not use heated lid).
- Add the following to each reaction from step 1:
| H2O | 4.5ul |
| Diluted SR RT primer | 1ul |
| Previous reaction product | 20ul |
| Total cumulative reaction volume | 25.5ul |
- Heat sample (3 different times and temperatures):
-
- 5 min at 750C (heated lid at 800C)
- 15 min at 370C (heated lid at 420C)
- 15 min at 250C (do not use heated lid). During this incubation, proceed to 'denature 5' SR adapter' below.
- Centrifuge to collect any condensation.
- Denature 5' SR adapter: With 5 minutes remaining in step C above, aliquot 1.1n ul of the 5’ adaptor into a PCR strip tube (where n = number of samples being processed).
- Incubate the 5’ adapter in a preheated thermal cycler at 700C for 2 minutes.
- Immediately place the tube on ice and use within 30 minutes.
- Add the following, individually, to the prepared RNA from steps above:
| Diluted 5’ SR adapter (denatured as above) | 1ul |
| 5’ ligation reaction buffer (10x) | 1ul |
| 5’ ligation enzyme mix |
2.5ul |
| Previous reaction product | 25.5ul |
| Total cumulative reaction volume | 30ul |
- Seal, flick thoroughly to mix, collect by centrifugation.
- Incubate for 1 hour at 250C (do not use heated lid).
- Make a stock of buffer, RNase inhibitor and RTase and use it to add the following to each rxn:
| First strand synthesis buffer | 8ul |
| Murine RNase inhibitor | 1ul |
| M-MuLV Reverse Transcriptase (RNase H-) | 1ul |
| Previous reaction product | 30ul |
| Total cumulative reaction volume | 40ul |
- Incubate for 1 hour at 500C.
- Cool on ice, collect condensation by centrifugation.
- Add the following to the RT reaction mix from step 4 (make a master mix of LongAmp Taq mix, diluted SR primer, and nuclease free water; add the Index primer individually).
| LongAmp Taq 2x Master mix | 50ul |
| Diluted SR primer | 2.5ul |
| Nuclease free water | 5.0ul |
| Inidividually added Index primer | 2.5ul |
| Previous reaction product | 40ul |
| Total volume | 100ul |
- Mix thoroughly by pipetting
- In some PCR cyclers, it may be necessary to split the reaction volume in to two separate 50ul reactions for efficient heating/cooling.
| Cycle Step |
TEMP |
Time |
Cycles |
| Initial denaturation |
940 C |
30 sec |
1 |
| Denaturation |
940 C |
15 sec |
12-15 |
| Annealing |
620 C |
30 sec |
12-15 |
| Extension |
700 C |
15 sec |
12-15 |
| Final Extension |
700 C |
5 min |
1 |
| Hold |
40 C |
- |
|
- Ethanol precipitate as above (pellet should be large).
- Dissolve the pellet in 20ul of TE buffer or ddH20.
- QC: Qubit (check ng/ul)
- Load 200ng of finished library into each well of a 4% EX E-gel, with 200ng of 50bp ladder as reference
<
- Run until smear is obvious from 150bp-300bp+, and products at 150bp can be easily resolved from any undesirable amplicons (common contaminant runs at 130bp)
>
- Run until smear is obvious from 200bp+, and products can be easily resolved from any undesirable amplicons at lower sizes.
- Take picture of gel with imager
- Open gel
- Excise bands from 150bp to the top of the ladder using a clean razor blade for each lane.
- Gel purify using Zymo gel recovery kit (use 550 C during gel dissolution step).
- Elute in 50-100ul H2O.
- Qubit for DNA concentration, submit to GSAF as per their recommendations.
<
- Example_RNAseq_libraries:
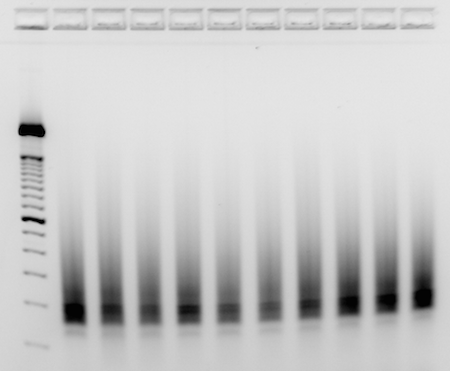
>
- Example_RNAseq_libraries:

| META FILEATTACHMENT | attachment="Barrick_Lab_2016-01-27_RNAlib_2.png" attr="" comment="Example_RNAseq_library" date="1485804500" name="Barrick_Lab_2016-01-27_RNAlib_2.png" path="Barrick Lab 2016-01-27 RNAlib_2.png" size="135027" stream="Barrick Lab 2016-01-27 RNAlib_2.png" tmpFilename="/usr/tmp/CGItemp24272" user="SimonDAlton" version="1" |
|---|
Revision 42017-01-30 - JeffreyBarrick
RNAseq Library PreparationUse RNA from RNASnap and Zymo coumn purification as outlined on wiki.rRNA DepletionUse the Ribozero (RZ) rRNA Removal Kit (Gram-negative bacteria) from Illumina. We assayed the effectiveness of the RZ kit: using inputs of up to 5ug, rRNA was reduced from >80% of total cell RNA to less than 1%. Protocol as follows: *Cool a benchtop centrifuge to 40C, place 100% ethanol and 70% ethanol on ice (store chilled at -200C). Set a waterbath to 500C* DO NOT PUT MAGNETIC BEADS ON ICE. All RZ components can be kept at room temperature during the procedure. Protocol: Batch wash magnetic beads:
ETOH precipitation purificationThe pellets - including the final pellet - may not be easily visible, so keep track of which 'corner' of the centrifuge tube is facing outwards in centrifuge. Protocol:
RNA FragmentationUse NEB Fragmentation Buffer. The RNA frag buffer conditions and high temperature cause RNA fragmentation. Fragment size is determined by time exposed to both. For tight control of fragmentation, add the buffer to the RNA in a strip tube, seal, resuspend tube by thoroughly flicking strip, centrifuge briefly and place in a pre-heated PCR cycler. Likewise, at the end of the run immediately cool on ice, collect any condensation by centrifugation and then add Stop Buffer. For E.coli, in our hands, 2 minutes at 94 degrees produces a final PCR product smear (see final steps) of between 150bp and 500bp (>98% of product between 150bp and 300bp). This should be determined experimentally from lab to lab. Protocol:
Kinase Treatment of Fragmented mRNAA master mix of buffer, T4 PNK and ATP can be made and 6ul added to n empty PCR strip tube wells (on ice) during the above ethanol precipitation's final 5 minute drying phase (see note above). Then resuspended RNA can be added to the reaction mix and thoroughly mixed immediately. Protocol:
Library Preparation
Use the NEBNext Small RNA Library Prep Set for Illumina (Multiplex Compatible), catalogue #E7330S/L (24/96 rxns).
The entire protocol is carried out in a single tube per sample, each subsequent reaction mixture being added to the previous product. Care should therefore be taken after each step to cool the products on ice and collect them by brief centrifugation so as to maintain reaction volume.
Note that many of the below steps require pre-heated blocks or transfers between temperatures. Plan accordingly.
Due to the low concentration of RNA following RZ, we routinely dilute the adapters, primers etc that come in the kit 1 in 2 (e.g. 1ul H2O + 1ul primer)
Protocol:
3’ SR Adaptor Ligation | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3' ligation buffer | 10ul |
| 3' ligation enzyme mix | 3ul |
| Total cumulative reaction volume | 20ul |
- Seal, flick thoroughly to mix, collect by centrifugation.
- Incubate for 1 hour at 250C (do not use heated lid).
- Add the following to each reaction from step 1:
| H2O | 4.5ul |
| Diluted SR RT primer | 1ul |
| Previous reaction product | 20ul |
| Total cumulative reaction volume | 25.5ul |
- Heat sample (3 different times and temperatures):
-
- 5 min at 750C (heated lid at 800C)
- 15 min at 370C (heated lid at 420C)
- 15 min at 250C (do not use heated lid). During this incubation, proceed to 'denature 5' SR adapter' below.
- Centrifuge to collect any condensation.
- Denature 5' SR adapter: With 5 minutes remaining in step C above, aliquot 1.1n ul of the 5’ adaptor into a PCR strip tube (where n = number of samples being processed).
- Incubate the 5’ adapter in a preheated thermal cycler at 700C for 2 minutes.
- Immediately place the tube on ice and use within 30 minutes.
- Add the following, individually, to the prepared RNA from steps above:
| Diluted 5’ SR adapter (denatured as above) | 1ul |
| 5’ ligation reaction buffer (10x) | 1ul |
| 5’ ligation enzyme mix |
2.5ul |
| Previous reaction product | 25.5ul |
| Total cumulative reaction volume | 30ul |
- Seal, flick thoroughly to mix, collect by centrifugation.
- Incubate for 1 hour at 250C (do not use heated lid).
- Make a stock of buffer, RNase inhibitor and RTase and use it to add the following to each rxn:
| First strand synthesis buffer | 8ul |
| Murine RNase inhibitor | 1ul |
| M-MuLV Reverse Transcriptase (RNase H-) | 1ul |
| Previous reaction product | 30ul |
| Total cumulative reaction volume | 40ul |
- Incubate for 1 hour at 500C.
- Cool on ice, collect condensation by centrifugation.
- Add the following to the RT reaction mix from step 4 (make a master mix of LongAmp Taq mix, diluted SR primer, and nuclease free water; add the Index primer individually).
| LongAmp Taq 2x Master mix | 50ul |
| Diluted SR primer | 2.5ul |
| Nuclease free water | 5.0ul |
| Inidividually added Index primer | 2.5ul |
| Previous reaction product | 40ul |
| Total volume | 100ul |
- Mix thoroughly by pipetting
- In some PCR cyclers, it may be necessary to split the reaction volume in to two separate 50ul reactions for efficient heating/cooling.
| Cycle Step |
TEMP |
Time |
Cycles |
| Initial denaturation |
940 C |
30 sec |
1 |
| Denaturation |
940 C |
15 sec |
12-15 |
| Annealing |
620 C |
30 sec |
12-15 |
| Extension |
700 C |
15 sec |
12-15 |
| Final Extension |
700 C |
5 min |
1 |
| Hold |
40 C |
- |
|
- Ethanol precipitate as above (pellet should be large).
- Dissolve the pellet in 20ul of TE buffer or ddH20.
- QC: Qubit (check ng/ul)
- Load 200ng of finished library into each well of a 4% EX E-gel, with 200ng of 50bp ladder as reference
- Run until smear is obvious from 150bp-300bp+, and products at 150bp can be easily resolved from any undesirable amplicons (common contaminant runs at 130bp)
- Take picture of gel with imager
- Open gel
- Excise bands from 150bp to the top of the ladder using a clean razor blade for each lane.
- Gel purify using Zymo gel recovery kit (use 550 C during gel dissolution step).
- Elute in 50-100ul H2O.
- Qubit for DNA concentration, submit to GSAF as per their recommendations.
- Example_RNAseq_libraries:

<
| META FILEATTACHMENT | attachment="Example_RNAseq_library.pdf" attr="" comment="Example_RNAseq_libraries" date="1485800884" name="Example_RNAseq_library.pdf" path="Barrick Lab 2016-01-27 RNAlib211-220,268.tif" size="2686320" stream="Barrick Lab 2016-01-27 RNAlib211-220,268.tif" tmpFilename="/usr/tmp/CGItemp29367" user="SimonDAlton" version="2" |
|---|---|
| META FILEATTACHMENT | attachment="Barrick_Lab_2016-01-27_RNAlib.png" attr="" comment="Example_library_prep" date="1485804310" name="Barrick_Lab_2016-01-27_RNAlib.png" path="Barrick Lab 2016-01-27 RNAlib.png" size="135027" stream="Barrick Lab 2016-01-27 RNAlib.png" tmpFilename="/usr/tmp/CGItemp26533" user="SimonDAlton" version="1" |
| META FILEATTACHMENT | attachment="Barrick_Lab_2016-01-27_RNAlib_2.png" attr="" comment="Example_RNAseq_library" date="1485804500" name="Barrick_Lab_2016-01-27_RNAlib_2.png" path="Barrick Lab 2016-01-27 RNAlib_2.png" size="135027" stream="Barrick Lab 2016-01-27 RNAlib_2.png" tmpFilename="/usr/tmp/CGItemp24272" user="SimonDAlton" version="1" |
|---|
Revision 32017-01-30 - SimonDAlton
RNAseq Library PreparationUse RNA from RNASnap and Zymo coumn purification as outlined on wiki.rRNA DepletionUse the Ribozero (RZ) rRNA Removal Kit (Gram-negative bacteria) from Illumina. We assayed the effectiveness of the RZ kit: using inputs of up to 5ug, rRNA was reduced from >80% of total cell RNA to less than 1%. Protocol as follows: *Cool a benchtop centrifuge to 40C, place 100% ethanol and 70% ethanol on ice (store chilled at -200C). Set a waterbath to 500C* DO NOT PUT MAGNETIC BEADS ON ICE. All RZ components can be kept at room temperature during the procedure. Protocol: Batch wash magnetic beads:
ETOH precipitation purificationThe pellets - including the final pellet - may not be easily visible, so keep track of which 'corner' of the centrifuge tube is facing outwards in centrifuge. Protocol:
RNA FragmentationUse NEB Fragmentation Buffer. The RNA frag buffer conditions and high temperature cause RNA fragmentation. Fragment size is determined by time exposed to both. For tight control of fragmentation, add the buffer to the RNA in a strip tube, seal, resuspend tube by thoroughly flicking strip, centrifuge briefly and place in a pre-heated PCR cycler. Likewise, at the end of the run immediately cool on ice, collect any condensation by centrifugation and then add Stop Buffer. For E.coli, in our hands, 2 minutes at 94 degrees produces a final PCR product smear (see final steps) of between 150bp and 500bp (>98% of product between 150bp and 300bp). This should be determined experimentally from lab to lab. Protocol:
Kinase Treatment of Fragmented mRNAA master mix of buffer, T4 PNK and ATP can be made and 6ul added to n empty PCR strip tube wells (on ice) during the above ethanol precipitation's final 5 minute drying phase (see note above). Then resuspended RNA can be added to the reaction mix and thoroughly mixed immediately. Protocol:
Library Preparation
Use the NEBNext Small RNA Library Prep Set for Illumina (Multiplex Compatible), catalogue #E7330S/L (24/96 rxns).
The entire protocol is carried out in a single tube per sample, each subsequent reaction mixture being added to the previous product. Care should therefore be taken after each step to cool the products on ice and collect them by brief centrifugation so as to maintain reaction volume.
Note that many of the below steps require pre-heated blocks or transfers between temperatures. Plan accordingly.
Due to the low concentration of RNA following RZ, we routinely dilute the adapters, primers etc that come in the kit 1 in 2 (e.g. 1ul H2O + 1ul primer)
Protocol:
3’ SR Adaptor Ligation | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3' ligation buffer | 10ul |
| 3' ligation enzyme mix | 3ul |
| Total cumulative reaction volume | 20ul |
- Seal, flick thoroughly to mix, collect by centrifugation.
- Incubate for 1 hour at 250C (do not use heated lid).
- Add the following to each reaction from step 1:
| H2O | 4.5ul |
| Diluted SR RT primer | 1ul |
| Previous reaction product | 20ul |
| Total cumulative reaction volume | 25.5ul |
- Heat sample (3 different times and temperatures):
-
- 5 min at 750C (heated lid at 800C)
- 15 min at 370C (heated lid at 420C)
- 15 min at 250C (do not use heated lid). During this incubation, proceed to 'denature 5' SR adapter' below.
- Centrifuge to collect any condensation.
- Denature 5' SR adapter: With 5 minutes remaining in step C above, aliquot 1.1n ul of the 5’ adaptor into a PCR strip tube (where n = number of samples being processed).
- Incubate the 5’ adapter in a preheated thermal cycler at 700C for 2 minutes.
- Immediately place the tube on ice and use within 30 minutes.
- Add the following, individually, to the prepared RNA from steps above:
| Diluted 5’ SR adapter (denatured as above) | 1ul |
| 5’ ligation reaction buffer (10x) | 1ul |
| 5’ ligation enzyme mix |
2.5ul |
| Previous reaction product | 25.5ul |
| Total cumulative reaction volume | 30ul |
- Seal, flick thoroughly to mix, collect by centrifugation.
- Incubate for 1 hour at 250C (do not use heated lid).
- Make a stock of buffer, RNase inhibitor and RTase and use it to add the following to each rxn:
| First strand synthesis buffer | 8ul |
| Murine RNase inhibitor | 1ul |
| M-MuLV Reverse Transcriptase (RNase H-) | 1ul |
| Previous reaction product | 30ul |
| Total cumulative reaction volume | 40ul |
- Incubate for 1 hour at 500C.
- Cool on ice, collect condensation by centrifugation.
- Add the following to the RT reaction mix from step 4 (make a master mix of LongAmp Taq mix, diluted SR primer, and nuclease free water; add the Index primer individually).
| LongAmp Taq 2x Master mix | 50ul |
| Diluted SR primer | 2.5ul |
| Nuclease free water | 5.0ul |
| Inidividually added Index primer | 2.5ul |
| Previous reaction product | 40ul |
| Total volume | 100ul |
- Mix thoroughly by pipetting
- In some PCR cyclers, it may be necessary to split the reaction volume in to two separate 50ul reactions for efficient heating/cooling.
| Cycle Step |
TEMP |
Time |
Cycles |
| Initial denaturation |
940 C |
30 sec |
1 |
| Denaturation |
940 C |
15 sec |
12-15 |
| Annealing |
620 C |
30 sec |
12-15 |
| Extension |
700 C |
15 sec |
12-15 |
| Final Extension |
700 C |
5 min |
1 |
| Hold |
40 C |
- |
|
- Ethanol precipitate as above (pellet should be large).
- Dissolve the pellet in 20ul of TE buffer or ddH20.
- QC: Qubit (check ng/ul)
- Load 200ng of finished library into each well of a 4% EX E-gel, with 200ng of 50bp ladder as reference
- Run until smear is obvious from 150bp-300bp+, and products at 150bp can be easily resolved from any undesirable amplicons (common contaminant runs at 130bp)
- Take picture of gel with imager
- Open gel
- Excise bands from 150bp to the top of the ladder using a clean razor blade for each lane.
- Gel purify using Zymo gel recovery kit (use 550 C during gel dissolution step).
- Elute in 50-100ul H2O.
- Qubit for DNA concentration, submit to GSAF as per their recommendations.
<
- Example_RNAseq_libraries:
>
- Example_RNAseq_libraries:

<

<
| META FILEATTACHMENT | attachment="Example_RNAseq_library.pdf" attr="" comment="Example_RNAseq_libraries" date="1485557879" name="Example_RNAseq_library.pdf" path="Example RNAseq library.pdf" size="1036154" stream="Example RNAseq library.pdf" tmpFilename="/usr/tmp/CGItemp24904" user="SimonDAlton" version="1" |
|---|
>
| META FILEATTACHMENT | attachment="Example_RNAseq_library.pdf" attr="" comment="Example_RNAseq_libraries" date="1485800884" name="Example_RNAseq_library.pdf" path="Barrick Lab 2016-01-27 RNAlib211-220,268.tif" size="2686320" stream="Barrick Lab 2016-01-27 RNAlib211-220,268.tif" tmpFilename="/usr/tmp/CGItemp29367" user="SimonDAlton" version="2" |
|---|
>
| META FILEATTACHMENT | attachment="Barrick_Lab_2016-01-27_RNAlib.png" attr="" comment="Example_library_prep" date="1485804310" name="Barrick_Lab_2016-01-27_RNAlib.png" path="Barrick Lab 2016-01-27 RNAlib.png" size="135027" stream="Barrick Lab 2016-01-27 RNAlib.png" tmpFilename="/usr/tmp/CGItemp26533" user="SimonDAlton" version="1" |
|---|---|
| META FILEATTACHMENT | attachment="Barrick_Lab_2016-01-27_RNAlib_2.png" attr="" comment="Example_RNAseq_library" date="1485804500" name="Barrick_Lab_2016-01-27_RNAlib_2.png" path="Barrick Lab 2016-01-27 RNAlib_2.png" size="135027" stream="Barrick Lab 2016-01-27 RNAlib_2.png" tmpFilename="/usr/tmp/CGItemp24272" user="SimonDAlton" version="1" |
Revision 22017-01-27 - SimonDAlton
RNAseq Library PreparationUse RNA from RNASnap and Zymo coumn purification as outlined on wiki.rRNA DepletionUse the Ribozero (RZ) rRNA Removal Kit (Gram-negative bacteria) from Illumina. We assayed the effectiveness of the RZ kit: using inputs of up to 5ug, rRNA was reduced from >80% of total cell RNA to less than 1%. Protocol as follows: *Cool a benchtop centrifuge to 40C, place 100% ethanol and 70% ethanol on ice (store chilled at -200C). Set a waterbath to 500C* DO NOT PUT MAGNETIC BEADS ON ICE. All RZ components can be kept at room temperature during the procedure. Protocol: Batch wash magnetic beads:
ETOH precipitation purificationThe pellets - including the final pellet - may not be easily visible, so keep track of which 'corner' of the centrifuge tube is facing outwards in centrifuge. Protocol:
RNA FragmentationUse NEB Fragmentation Buffer. The RNA frag buffer conditions and high temperature cause RNA fragmentation. Fragment size is determined by time exposed to both. For tight control of fragmentation, add the buffer to the RNA in a strip tube, seal, resuspend tube by thoroughly flicking strip, centrifuge briefly and place in a pre-heated PCR cycler. Likewise, at the end of the run immediately cool on ice, collect any condensation by centrifugation and then add Stop Buffer. For E.coli, in our hands, 2 minutes at 94 degrees produces a final PCR product smear (see final steps) of between 150bp and 500bp (>98% of product between 150bp and 300bp). This should be determined experimentally from lab to lab. Protocol:
Kinase Treatment of Fragmented mRNAA master mix of buffer, T4 PNK and ATP can be made and 6ul added to n empty PCR strip tube wells (on ice) during the above ethanol precipitation's final 5 minute drying phase (see note above). Then resuspended RNA can be added to the reaction mix and thoroughly mixed immediately. Protocol:
Library Preparation
Use the NEBNext Small RNA Library Prep Set for Illumina (Multiplex Compatible), catalogue #E7330S/L (24/96 rxns).
The entire protocol is carried out in a single tube per sample, each subsequent reaction mixture being added to the previous product. Care should therefore be taken after each step to cool the products on ice and collect them by brief centrifugation so as to maintain reaction volume.
Note that many of the below steps require pre-heated blocks or transfers between temperatures. Plan accordingly.
Due to the low concentration of RNA following RZ, we routinely dilute the adapters, primers etc that come in the kit 1 in 2 (e.g. 1ul H2O + 1ul primer)
Protocol:
3’ SR Adaptor Ligation | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| 3' ligation buffer | 10ul |
| 3' ligation enzyme mix | 3ul |
| Total cumulative reaction volume | 20ul |
- Seal, flick thoroughly to mix, collect by centrifugation.
- Incubate for 1 hour at 250C (do not use heated lid).
- Add the following to each reaction from step 1:
| H2O | 4.5ul |
| Diluted SR RT primer | 1ul |
| Previous reaction product | 20ul |
| Total cumulative reaction volume | 25.5ul |
- Heat sample (3 different times and temperatures):
-
- 5 min at 750C (heated lid at 800C)
- 15 min at 370C (heated lid at 420C)
- 15 min at 250C (do not use heated lid). During this incubation, proceed to 'denature 5' SR adapter' below.
- Centrifuge to collect any condensation.
- Denature 5' SR adapter: With 5 minutes remaining in step C above, aliquot 1.1n ul of the 5’ adaptor into a PCR strip tube (where n = number of samples being processed).
- Incubate the 5’ adapter in a preheated thermal cycler at 700C for 2 minutes.
- Immediately place the tube on ice and use within 30 minutes.
- Add the following, individually, to the prepared RNA from steps above:
| Diluted 5’ SR adapter (denatured as above) | 1ul |
| 5’ ligation reaction buffer (10x) | 1ul |
| 5’ ligation enzyme mix |
2.5ul |
| Previous reaction product | 25.5ul |
| Total cumulative reaction volume | 30ul |
- Seal, flick thoroughly to mix, collect by centrifugation.
- Incubate for 1 hour at 250C (do not use heated lid).
- Make a stock of buffer, RNase inhibitor and RTase and use it to add the following to each rxn:
| First strand synthesis buffer | 8ul |
| Murine RNase inhibitor | 1ul |
| M-MuLV Reverse Transcriptase (RNase H-) | 1ul |
| Previous reaction product | 30ul |
| Total cumulative reaction volume | 40ul |
- Incubate for 1 hour at 500C.
- Cool on ice, collect condensation by centrifugation.
- Add the following to the RT reaction mix from step 4 (make a master mix of LongAmp Taq mix, diluted SR primer, and nuclease free water; add the Index primer individually).
| LongAmp Taq 2x Master mix | 50ul |
| Diluted SR primer | 2.5ul |
| Nuclease free water | 5.0ul |
| Inidividually added Index primer | 2.5ul |
| Previous reaction product | 40ul |
| Total volume | 100ul |
- Mix thoroughly by pipetting
- In some PCR cyclers, it may be necessary to split the reaction volume in to two separate 50ul reactions for efficient heating/cooling.
| Cycle Step |
TEMP |
Time |
Cycles |
| Initial denaturation |
940 C |
30 sec |
1 |
| Denaturation |
940 C |
15 sec |
12-15 |
| Annealing |
620 C |
30 sec |
12-15 |
| Extension |
700 C |
15 sec |
12-15 |
| Final Extension |
700 C |
5 min |
1 |
| Hold |
40 C |
- |
|
- Ethanol precipitate as above (pellet should be large).
- Dissolve the pellet in 20ul of TE buffer or ddH20.
- QC: Qubit (check ng/ul)
- Load 200ng of finished library into each well of a 4% EX E-gel, with 200ng of 50bp ladder as reference
- Run until smear is obvious from 150bp-300bp+, and products at 150bp can be easily resolved from any undesirable amplicons (common contaminant runs at 130bp)
- Take picture of gel with imager
- Open gel
- Excise bands from 150bp to the top of the ladder using a clean razor blade for each lane.
- Gel purify using Zymo gel recovery kit (use 550 C during gel dissolution step).
- Elute in 50-100ul H2O.
- Qubit for DNA concentration, submit to GSAF as per their recommendations.
<
>
- Example_RNAseq_libraries:
>

| META FILEATTACHMENT | attachment="Example_RNAseq_library.pdf" attr="" comment="Example_RNAseq_libraries" date="1485557879" name="Example_RNAseq_library.pdf" path="Example RNAseq library.pdf" size="1036154" stream="Example RNAseq library.pdf" tmpFilename="/usr/tmp/CGItemp24904" user="SimonDAlton" version="1" |
|---|
Revision 12017-01-27 - SimonDAlton
RNAseq Library PreparationUse RNA from RNASnap and Zymo coumn purification as outlined on wiki.rRNA DepletionUse the Ribozero (RZ) rRNA Removal Kit (Gram-negative bacteria) from Illumina. We assayed the effectiveness of the RZ kit: using inputs of up to 5ug, rRNA was reduced from >80% of total cell RNA to less than 1%. Protocol as follows: *Cool a benchtop centrifuge to 40C, place 100% ethanol and 70% ethanol on ice (store chilled at -200C). Set a waterbath to 500C* DO NOT PUT MAGNETIC BEADS ON ICE. All RZ components can be kept at room temperature during the procedure. Protocol: Batch wash magnetic beads:
ETOH precipitation purificationThe pellets - including the final pellet - may not be easily visible, so keep track of which 'corner' of the centrifuge tube is facing outwards in centrifuge. Protocol:
RNA FragmentationUse NEB Fragmentation Buffer. The RNA frag buffer conditions and high temperature cause RNA fragmentation. Fragment size is determined by time exposed to both. For tight control of fragmentation, add the buffer to the RNA in a strip tube, seal, resuspend tube by thoroughly flicking strip, centrifuge briefly and place in a pre-heated PCR cycler. Likewise, at the end of the run immediately cool on ice, collect any condensation by centrifugation and then add Stop Buffer. For E.coli, in our hands, 2 minutes at 94 degrees produces a final PCR product smear (see final steps) of between 150bp and 500bp (>98% of product between 150bp and 300bp). This should be determined experimentally from lab to lab. Protocol:
Kinase Treatment of Fragmented mRNAA master mix of buffer, T4 PNK and ATP can be made and 6ul added to n empty PCR strip tube wells (on ice) during the above ethanol precipitation's final 5 minute drying phase (see note above). Then resuspended RNA can be added to the reaction mix and thoroughly mixed immediately. Protocol:
Library Preparation
Use the NEBNext Small RNA Library Prep Set for Illumina (Multiplex Compatible), catalogue #E7330S/L (24/96 rxns).
The entire protocol is carried out in a single tube per sample, each subsequent reaction mixture being added to the previous product. Care should therefore be taken after each step to cool the products on ice and collect them by brief centrifugation so as to maintain reaction volume.
Note that many of the below steps require pre-heated blocks or transfers between temperatures. Plan accordingly.
Due to the low concentration of RNA following RZ, we routinely dilute the adapters, primers etc that come in the kit 1 in 2 (e.g. 1ul H2O + 1ul primer)
Protocol:
3’ SR Adaptor Ligation |
| 3' ligation buffer | 10ul |
| 3' ligation enzyme mix | 3ul |
| Total cumulative reaction volume | 20ul |
- Seal, flick thoroughly to mix, collect by centrifugation.
- Incubate for 1 hour at 250C (do not use heated lid).
- Add the following to each reaction from step 1:
| H2O | 4.5ul |
| Diluted SR RT primer | 1ul |
| Previous reaction product | 20ul |
| Total cumulative reaction volume | 25.5ul |
- Heat sample (3 different times and temperatures):
-
- 5 min at 750C (heated lid at 800C)
- 15 min at 370C (heated lid at 420C)
- 15 min at 250C (do not use heated lid). During this incubation, proceed to 'denature 5' SR adapter' below.
- Centrifuge to collect any condensation.
- Denature 5' SR adapter: With 5 minutes remaining in step C above, aliquot 1.1n ul of the 5’ adaptor into a PCR strip tube (where n = number of samples being processed).
- Incubate the 5’ adapter in a preheated thermal cycler at 700C for 2 minutes.
- Immediately place the tube on ice and use within 30 minutes.
- Add the following, individually, to the prepared RNA from steps above:
| Diluted 5’ SR adapter (denatured as above) | 1ul |
| 5’ ligation reaction buffer (10x) | 1ul |
| 5’ ligation enzyme mix |
2.5ul |
| Previous reaction product | 25.5ul |
| Total cumulative reaction volume | 30ul |
- Seal, flick thoroughly to mix, collect by centrifugation.
- Incubate for 1 hour at 250C (do not use heated lid).
- Make a stock of buffer, RNase inhibitor and RTase and use it to add the following to each rxn:
| First strand synthesis buffer | 8ul |
| Murine RNase inhibitor | 1ul |
| M-MuLV Reverse Transcriptase (RNase H-) | 1ul |
| Previous reaction product | 30ul |
| Total cumulative reaction volume | 40ul |
- Incubate for 1 hour at 500C.
- Cool on ice, collect condensation by centrifugation.
- Add the following to the RT reaction mix from step 4 (make a master mix of LongAmp Taq mix, diluted SR primer, and nuclease free water; add the Index primer individually).
| LongAmp Taq 2x Master mix | 50ul |
| Diluted SR primer | 2.5ul |
| Nuclease free water | 5.0ul |
| Inidividually added Index primer | 2.5ul |
| Previous reaction product | 40ul |
| Total volume | 100ul |
- Mix thoroughly by pipetting
- In some PCR cyclers, it may be necessary to split the reaction volume in to two separate 50ul reactions for efficient heating/cooling.
| Cycle Step |
TEMP |
Time |
Cycles |
| Initial denaturation |
940 C |
30 sec |
1 |
| Denaturation |
940 C |
15 sec |
12-15 |
| Annealing |
620 C |
30 sec |
12-15 |
| Extension |
700 C |
15 sec |
12-15 |
| Final Extension |
700 C |
5 min |
1 |
| Hold |
40 C |
- |
|
- Ethanol precipitate as above (pellet should be large).
- Dissolve the pellet in 20ul of TE buffer or ddH20.
- QC: Qubit (check ng/ul)
- Load 200ng of finished library into each well of a 4% EX E-gel, with 200ng of 50bp ladder as reference
- Run until smear is obvious from 150bp-300bp+, and products at 150bp can be easily resolved from any undesirable amplicons (common contaminant runs at 130bp)
- Take picture of gel with imager
- Open gel
- Excise bands from 150bp to the top of the ladder using a clean razor blade for each lane.
- Gel purify using Zymo gel recovery kit (use 550 C during gel dissolution step).
- Elute in 50-100ul H2O.
- Qubit for DNA concentration, submit to GSAF as per their recommendations.