Difference: ProtocolsGibsonCloning (1 vs. 16)
Revision 162021-10-22 - CameronRoots
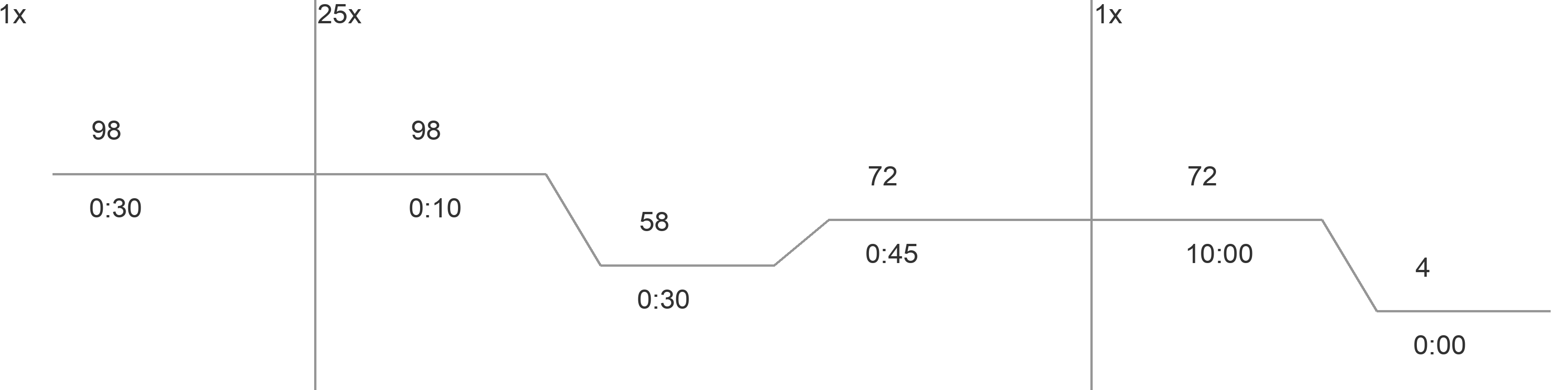
Gibson AssemblyBackground and DesignGibson Cloning is a technique of DNA construct assembly that allows one to join multiple linear segments into either one large linear segment or, if the segments contain the appropriate components and overlaps, an intact plasmid. This protocol follows the one-step isothermal assembly of overlapping dsDNA. In order to assemble segments of DNA via Gibson Cloning, they usually must contain at least 20bp of homology to the segment they are being joined to (Tm of overlapping region must be >= 48°C). Homology overlaps can vary in length from as few as 15bps up to 80bps -- efficacy depends on number of fragments assembled, as well as brand of "Gibson Mastermix" used. ie NEBuilder Hi-Fi DNA Assembly Mix will have a different optimal overlap length than NEB Gibson Master Mix and a different optimal length for homebrew Gibson Master Mix (recipe attached at bottom). Generally, 20bp overlap with proper a Tm is suitable. For example if one was to make a construct that was Seg1-Seg2-Seg3 from individual PCRs of Seg1, Seg2, and Seg3 (as indicated in the figure below) the 3' end of Seg1 would need at least 20bp of homology to the 5' end of Seg2 and Seg3 would require its 5' end to have at least 20bp of homology to Seg2. For constructing a plasmid, use a linearized vector backbone as one of your segments.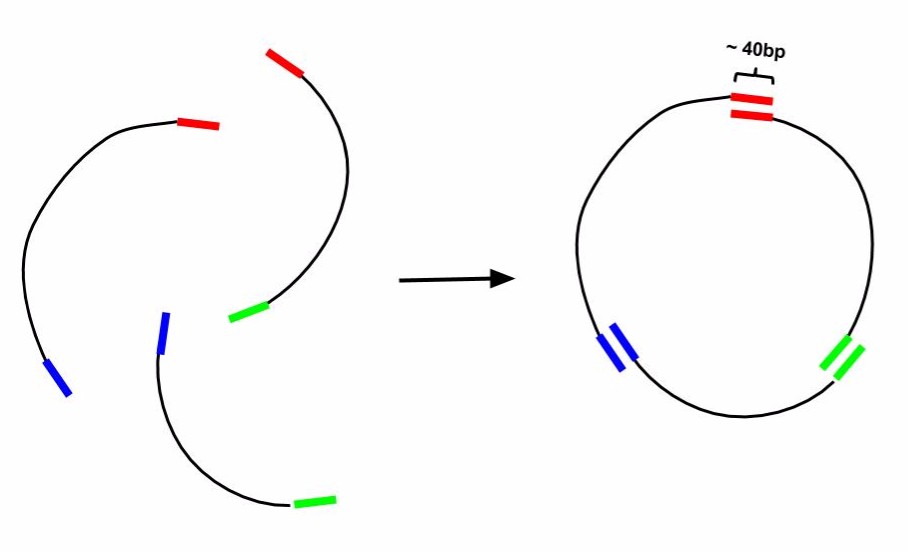
SuppliesIf homemade master mix aliquots are available, and less than 1 year old:
Primer Design Using GibsonFor a thorough discussion on the construction of primers for use in Gibson Assembly, please see the following publication: http://www.ncbi.nlm.nih.gov/pubmed/21601685.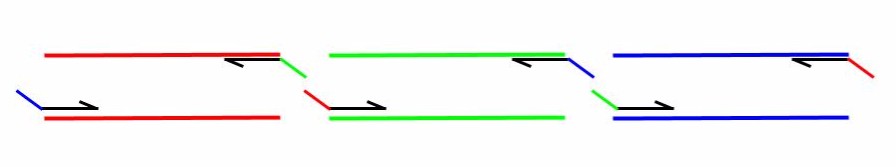 A note on primer design = Try to design your junctions to be in between the origin of replication or the selectable marker on the plasmid that you want to make. This will reduce your number of false-positives.
A note on primer design = Try to design your junctions to be in between the origin of replication or the selectable marker on the plasmid that you want to make. This will reduce your number of false-positives.
Protocol
Expected ResultsThe positive control should yield ampicillin resistant colonies when transformed into E. coli. You will see different efficiencies depending on the number of fragments in your reaction. Gibson reactions with a higher number of fragments tend to be less efficient, therefore you will likely see few colonies on your plates.Positive Control Construction and UsageThe Positive Control DNA Mix for Gibson Assembly consists of a two-piece assembly of pUC19. It is designed such that 5uL of the Positive Control DNA Mix is to be added to 15uL of Gibson Assembly Master Mix along side experimental reactions. Both pUC19 segments are between 1.3kb and 1.4kb in size. To construct the positive control reaction mix:
| |||||||||||
| Added: | |||||||||||
| > > |
| ||||||||||
| |||||||||||
| Added: | |||||||||||
| > > |
| ||||||||||
Revision 152018-03-22 - DaciaLeon
Gibson AssemblyBackground and DesignGibson Cloning is a technique of DNA construct assembly that allows one to join multiple linear segments into either one large linear segment or, if the segments contain the appropriate components and overlaps, an intact plasmid. This protocol follows the one-step isothermal assembly of overlapping dsDNA. In order to assemble segments of DNA via Gibson Cloning, they usually must contain at least 20bp of homology to the segment they are being joined to (Tm of overlapping region must be >= 48°C). Homology overlaps can vary in length from as few as 15bps up to 80bps -- efficacy depends on number of fragments assembled, as well as brand of "Gibson Mastermix" used. ie NEBuilder Hi-Fi DNA Assembly Mix will have a different optimal overlap length than NEB Gibson Master Mix and a different optimal length for homebrew Gibson Master Mix (recipe attached at bottom). Generally, 20bp overlap with proper a Tm is suitable. For example if one was to make a construct that was Seg1-Seg2-Seg3 from individual PCRs of Seg1, Seg2, and Seg3 (as indicated in the figure below) the 3' end of Seg1 would need at least 20bp of homology to the 5' end of Seg2 and Seg3 would require its 5' end to have at least 20bp of homology to Seg2. For constructing a plasmid, use a linearized vector backbone as one of your segments.
SuppliesIf homemade master mix aliquots are available, and less than 1 year old: | |||||||||||
| Changed: | |||||||||||
| < < |
| ||||||||||
| > > |
| ||||||||||
Primer Design Using GibsonFor a thorough discussion on the construction of primers for use in Gibson Assembly, please see the following publication: http://www.ncbi.nlm.nih.gov/pubmed/21601685. | |||||||||||
| Added: | |||||||||||
| > > | A note on primer design = Try to design your junctions to be in between the origin of replication or the selectable marker on the plasmid that you want to make. This will reduce your number of false-positives. | ||||||||||
Protocol | |||||||||||
| Changed: | |||||||||||
| < < |
| ||||||||||
| > > |
| ||||||||||
| Added: | |||||||||||
| > > |
| ||||||||||
Expected Results | |||||||||||
| Changed: | |||||||||||
| < < | The positive control should yield ampicillin resistant colonies when transformed into E. coli. | ||||||||||
| > > | The positive control should yield ampicillin resistant colonies when transformed into E. coli. You will see different efficiencies depending on the number of fragments in your reaction. Gibson reactions with a higher number of fragments tend to be less efficient, therefore you will likely see few colonies on your plates. | ||||||||||
Positive Control Construction and UsageThe Positive Control DNA Mix for Gibson Assembly consists of a two-piece assembly of pUC19. It is designed such that 5uL of the Positive Control DNA Mix is to be added to 15uL of Gibson Assembly Master Mix along side experimental reactions. Both pUC19 segments are between 1.3kb and 1.4kb in size. To construct the positive control reaction mix:
| |||||||||||
| Changed: | |||||||||||
| < < |
| ||||||||||
| > > |
| ||||||||||
| |||||||||||
| Changed: | |||||||||||
| < < |
| ||||||||||
| > > |
| ||||||||||
| |||||||||||
| Changed: | |||||||||||
| < < |
| ||||||||||
| > > |
| ||||||||||
| |||||||||||
| Changed: | |||||||||||
| < < |
| ||||||||||
| > > |
| ||||||||||
| Changed: | |||||||||||
| < < | Using 5uL of this reaction provides approximately 1ng/100bp of each fragment in the Gibson Assembly reaction. | ||||||||||
| > > | Using 5µL of this reaction provides approximately 1ng/100bp of each fragment in the Gibson Assembly reaction. | ||||||||||
| |||||||||||
Revision 142017-12-14 - MattMcGuffie
Gibson AssemblyBackground and Design | ||||||||
| Changed: | ||||||||
| < < |  | |||||||
| > > | Gibson Cloning is a technique of DNA construct assembly that allows one to join multiple linear segments into either one large linear segment or, if the segments contain the appropriate components and overlaps, an intact plasmid. | |||||||
| Deleted: | ||||||||
| < < | Gibson Cloning is a technique of DNA construct assembly that allows one to join multiple linear segments into either one large linear segment or, if the segments contain the appropriate components and overlaps, an intact plasmid. This protocol follows the one-step ISO assembly of overlapping dsDNA protocol. In order to assemble segments of DNA via Gibson Cloning, they must contain at least 40bp of homology to the segment they are being joined to. For example, if one was to make a construct that was Seg1-Seg2-Seg3 from individual PCRs of Seg1, Seg2, and Seg3, the 3' end of Seg1 would need 40bp of homology to the 5' end of Seg2 and Seg3 would require its 5' end to have 40bp of homology to Seg2. For constructing a plasmid, use a linearized vector backbone as one of your segments. | |||||||
| Added: | ||||||||
| > > | This protocol follows the one-step isothermal assembly of overlapping dsDNA. In order to assemble segments of DNA via Gibson Cloning, they usually must contain at least 20bp of homology to the segment they are being joined to (Tm of overlapping region must be >= 48°C). Homology overlaps can vary in length from as few as 15bps up to 80bps -- efficacy depends on number of fragments assembled, as well as brand of "Gibson Mastermix" used. ie NEBuilder Hi-Fi DNA Assembly Mix will have a different optimal overlap length than NEB Gibson Master Mix and a different optimal length for homebrew Gibson Master Mix (recipe attached at bottom). Generally, 20bp overlap with proper a Tm is suitable.
For example if one was to make a construct that was Seg1-Seg2-Seg3 from individual PCRs of Seg1, Seg2, and Seg3 (as indicated in the figure below) the 3' end of Seg1 would need at least 20bp of homology to the 5' end of Seg2 and Seg3 would require its 5' end to have at least 20bp of homology to Seg2. For constructing a plasmid, use a linearized vector backbone as one of your segments.
 | |||||||
Supplies | ||||||||
| Changed: | ||||||||
| < < | If master mix aliquots are available, and less than 1 year old: | |||||||
| > > | If homemade master mix aliquots are available, and less than 1 year old: | |||||||
Primer Design Using Gibson | ||||||||
| Changed: | ||||||||
| < < | For a thorough discussion on the construction of primers for use in Gibson Assembly, please see the following publication: http://www.ncbi.nlm.nih.gov/pubmed/21601685. | |||||||
| > > | For a thorough discussion on the construction of primers for use in Gibson Assembly, please see the following publication: http://www.ncbi.nlm.nih.gov/pubmed/21601685.  | |||||||
| Deleted: | ||||||||
| < < |  | |||||||
Protocol
| ||||||||
| Changed: | ||||||||
| < < |
| |||||||
| > > |
| |||||||
Expected ResultsThe positive control should yield ampicillin resistant colonies when transformed into E. coli.Positive Control Construction and UsageThe Positive Control DNA Mix for Gibson Assembly consists of a two-piece assembly of pUC19. It is designed such that 5uL of the Positive Control DNA Mix is to be added to 15uL of Gibson Assembly Master Mix along side experimental reactions. Both pUC19 segments are between 1.3kb and 1.4kb in size. To construct the positive control reaction mix:
| ||||||||
| Added: | ||||||||
| > > | * | |||||||
| Added: | ||||||||
| > > |
| |||||||
| ||||||||
| Added: | ||||||||
| > > |
| |||||||
Revision 132017-09-05 - SimonDAlton
| ||||||||
| Changed: | ||||||||
| < < | Gibson Assembly | |||||||
| > > | Gibson Assembly | |||||||
| Changed: | ||||||||
| < < | Background and Design | |||||||
| > > | Background and Design | |||||||
| Deleted: | ||||||||
| < < | 
| |||||||
| Changed: | ||||||||
| < < | Supplies | |||||||
| > > |  | |||||||
| Added: | ||||||||
| > > | Gibson Cloning is a technique of DNA construct assembly that allows one to join multiple linear segments into either one large linear segment or, if the segments contain the appropriate components and overlaps, an intact plasmid. This protocol follows the one-step ISO assembly of overlapping dsDNA protocol. In order to assemble segments of DNA via Gibson Cloning, they must contain at least 40bp of homology to the segment they are being joined to. For example, if one was to make a construct that was Seg1-Seg2-Seg3 from individual PCRs of Seg1, Seg2, and Seg3, the 3' end of Seg1 would need 40bp of homology to the 5' end of Seg2 and Seg3 would require its 5' end to have 40bp of homology to Seg2. For constructing a plasmid, use a linearized vector backbone as one of your segments. | |||||||
| Added: | ||||||||
| > > | Supplies | |||||||
If master mix aliquots are available, and less than 1 year old:
| ||||||||
| Deleted: | ||||||||
| < < | If no master mix aliquots remain, but there is still <1 year old isothermal (ISO) reaction buffer available: | |||||||
| Added: | ||||||||
| > > | If master mix aliquots are not available, make more using guidelines in the attached document. | |||||||
| Added: | ||||||||
| > > | Primer Design Using Gibson | |||||||
| Deleted: | ||||||||
| < < |
If no master mix aliquots remain, and there is no ISO buffer, or the current ISO buffer is more than a year old:
Primer Design Using Gibson | |||||||
| For a thorough discussion on the construction of primers for use in Gibson Assembly, please see the following publication: http://www.ncbi.nlm.nih.gov/pubmed/21601685. | ||||||||
| Changed: | ||||||||
| < < |  | |||||||
| > > |  | |||||||
| Changed: | ||||||||
| < < | Protocol | |||||||
| > > | Protocol | |||||||
| ||||||||
| Changed: | ||||||||
| < < |
| |||||||
| > > |
| |||||||
| ||||||||
| Changed: | ||||||||
| < < |
| |||||||
| > > |
| |||||||
| Changed: | ||||||||
| < < | Expected Results | |||||||
| > > | Expected Results | |||||||
The positive control should yield ampicillin resistant colonies when transformed into E. coli.
Positive Control Construction and UsageThe Positive Control DNA Mix for Gibson Assembly consists of a two-piece assembly of pUC19. It is designed such that 5uL of the Positive Control DNA Mix is to be added to 15uL of Gibson Assembly Master Mix along side experimental reactions. Both pUC19 segments are between 1.3kb and 1.4kb in size. To construct the positive control reaction mix: | ||||||||
| Changed: | ||||||||
| < < |
| |||||||
| > > |
| |||||||
| ||||||||
| Changed: | ||||||||
| < < |
| |||||||
| > > |
| |||||||
| ||||||||
| Changed: | ||||||||
| < < |
| |||||||
| > > |
| |||||||
| ||||||||
| Changed: | ||||||||
| < < |
| |||||||
| > > |
| |||||||
| ||||||||
| Changed: | ||||||||
| < < |
| |||||||
| > > |
| |||||||
| Changed: | ||||||||
| < < | Using 5uL of this reaction provides approximately 1ng/100bp of each fragment in the Gibson Assembly reaction. | |||||||
| > > | Using 5uL of this reaction provides approximately 1ng/100bp of each fragment in the Gibson Assembly reaction. | |||||||
| Added: | ||||||||
| > > |
| |||||||
| ||||||||
| Added: | ||||||||
| > > |
| |||||||
Revision 122016-12-16 - SimonDOelsnitz
Gibson AssemblyBackground and Design | ||||||||
| Added: | ||||||||
| > > | 
| |||||||
| Deleted: | ||||||||
| < < | Gibson Cloning is a technique of DNA construct assembly that allows one to join multiple linear segments into either one large linear segment or, if the segments contain the appropriate components and overlaps, an intact plasmid. This protocol follows the one-step ISO assembly of overlapping dsDNA protocol. In order to assemble segments of DNA via Gibson Cloning, they must contain at least 40bp of homology to the segment they are being joined to. For example, if one was to make a construct that was Seg1-Seg2-Seg3 from individual PCRs of Seg1, Seg2, and Seg3, the 3' end of Seg1 would need 40bp of homology to the 5' end of Seg2 and Seg3 would require it 5' end to have 40bp of homology to Seg2. For constructing a plasmid, use a linearized vector backbone as one of your segments.
 | |||||||
SuppliesIf master mix aliquots are available, and less than 1 year old: | ||||||||
| Changed: | ||||||||
| < < |
| |||||||
| > > |
| |||||||
Primer Design Using GibsonFor a thorough discussion on the construction of primers for use in Gibson Assembly, please see the following publication: http://www.ncbi.nlm.nih.gov/pubmed/21601685. | ||||||||
| Added: | ||||||||
| > > |  | |||||||
Protocol
Expected ResultsThe positive control should yield ampicillin resistant colonies when transformed into E. coli.Positive Control Construction and UsageThe Positive Control DNA Mix for Gibson Assembly consists of a two-piece assembly of pUC19. It is designed such that 5uL of the Positive Control DNA Mix is to be added to 15uL of Gibson Assembly Master Mix along side experimental reactions. Both pUC19 segments are between 1.3kb and 1.4kb in size. To construct the positive control reaction mix:
| ||||||||
| Added: | ||||||||
| > > |
| |||||||
Revision 112016-12-16 - JeffreyBarrick
Gibson AssemblyBackground and DesignGibson Cloning is a technique of DNA construct assembly that allows one to join multiple linear segments into either one large linear segment or, if the segments contain the appropriate components and overlaps, an intact plasmid. This protocol follows the one-step ISO assembly of overlapping dsDNA protocol. In order to assemble segments of DNA via Gibson Cloning, they must contain at least 40bp of homology to the segment they are being joined to. For example, if one was to make a construct that was Seg1-Seg2-Seg3 from individual PCRs of Seg1, Seg2, and Seg3, the 3' end of Seg1 would need 40bp of homology to the 5' end of Seg2 and Seg3 would require it 5' end to have 40bp of homology to Seg2. For constructing a plasmid, use a linearized vector backbone as one of your segments.
SuppliesIf master mix aliquots are available, and less than 1 year old:
Primer Design Using GibsonFor a thorough discussion on the construction of primers for use in Gibson Assembly, please see the following publication: http://www.ncbi.nlm.nih.gov/pubmed/21601685.Protocol
Expected ResultsThe positive control should yield ampicillin resistant colonies when transformed into E. coli.Positive Control Construction and UsageThe Positive Control DNA Mix for Gibson Assembly consists of a two-piece assembly of pUC19. It is designed such that 5uL of the Positive Control DNA Mix is to be added to 15uL of Gibson Assembly Master Mix along side experimental reactions. Both pUC19 segments are between 1.3kb and 1.4kb in size. To construct the positive control reaction mix:
| ||||||||
| Changed: | ||||||||
| < < | -- Main.BrianRenda - 23 May 2014 | |||||||
| > > |
| |||||||
| Deleted: | ||||||||
| < < |
| |||||||
Revision 102016-12-16 - SimonDOelsnitz
Gibson AssemblyBackground and DesignGibson Cloning is a technique of DNA construct assembly that allows one to join multiple linear segments into either one large linear segment or, if the segments contain the appropriate components and overlaps, an intact plasmid. This protocol follows the one-step ISO assembly of overlapping dsDNA protocol. In order to assemble segments of DNA via Gibson Cloning, they must contain at least 40bp of homology to the segment they are being joined to. For example, if one was to make a construct that was Seg1-Seg2-Seg3 from individual PCRs of Seg1, Seg2, and Seg3, the 3' end of Seg1 would need 40bp of homology to the 5' end of Seg2 and Seg3 would require it 5' end to have 40bp of homology to Seg2. For constructing a plasmid, use a linearized vector backbone as one of your segments. | ||||||||
| Added: | ||||||||
| > > |
 | |||||||
SuppliesIf master mix aliquots are available, and less than 1 year old:
Primer Design Using GibsonFor a thorough discussion on the construction of primers for use in Gibson Assembly, please see the following publication: http://www.ncbi.nlm.nih.gov/pubmed/21601685.Protocol
Expected ResultsThe positive control should yield ampicillin resistant colonies when transformed into E. coli.Positive Control Construction and UsageThe Positive Control DNA Mix for Gibson Assembly consists of a two-piece assembly of pUC19. It is designed such that 5uL of the Positive Control DNA Mix is to be added to 15uL of Gibson Assembly Master Mix along side experimental reactions. Both pUC19 segments are between 1.3kb and 1.4kb in size. To construct the positive control reaction mix:
| ||||||||
| Added: | ||||||||
| > > |
| |||||||
Revision 92014-05-23 - BrianRenda
| ||||||||
| Changed: | ||||||||
| < < | Gibson Assembly-Work In Progress, do not use yet | |||||||
| > > | Gibson Assembly | |||||||
Background and DesignGibson Cloning is a technique of DNA construct assembly that allows one to join multiple linear segments into either one large linear segment or, if the segments contain the appropriate components and overlaps, an intact plasmid. This protocol follows the one-step ISO assembly of overlapping dsDNA protocol. In order to assemble segments of DNA via Gibson Cloning, they must contain at least 40bp of homology to the segment they are being joined to. For example, if one was to make a construct that was Seg1-Seg2-Seg3 from individual PCRs of Seg1, Seg2, and Seg3, the 3' end of Seg1 would need 40bp of homology to the 5' end of Seg2 and Seg3 would require it 5' end to have 40bp of homology to Seg2. For constructing a plasmid, use a linearized vector backbone as one of your segments.Supplies | ||||||||
| Changed: | ||||||||
| < < | If mastermix aliquots are available, and less than 1 year old: | |||||||
| > > | If master mix aliquots are available, and less than 1 year old: | |||||||
| ||||||||
| Changed: | ||||||||
| < < |
| |||||||
| > > |
| |||||||
| Added: | ||||||||
| > > | If no master mix aliquots remain, but there is still <1 year old isothermal (ISO) reaction buffer available: | |||||||
| Deleted: | ||||||||
| < < | If no mastermix aliquots remain, but there is still <1 year old isothermal (ISO) reaction buffer available: | |||||||
| Added: | ||||||||
| > > | If no master mix aliquots remain, and there is no ISO buffer, or the current ISO buffer is more than a year old: | |||||||
| Deleted: | ||||||||
| < < | If no mastermix aliquots remain, and there is no ISO buffer, or the current ISO buffer is more than a year old: | |||||||
| Added: | ||||||||
| > > | Primer Design Using Gibson | |||||||
| Changed: | ||||||||
| < < | Primer Design Using Gibthon | |||||||
| > > | For a thorough discussion on the construction of primers for use in Gibson Assembly, please see the following publication: http://www.ncbi.nlm.nih.gov/pubmed/21601685. | |||||||
| Deleted: | ||||||||
| < < | ||||||||
Protocol
Expected ResultsThe positive control should yield ampicillin resistant colonies when transformed into E. coli. | ||||||||
| Changed: | ||||||||
| < < | Positive control details | |||||||
| > > | Positive Control Construction and Usage | |||||||
| Changed: | ||||||||
| < < | PCR using these primer pairs and pUC19 as the DNA template: | |||||||
| > > | The Positive Control DNA Mix for Gibson Assembly consists of a two-piece assembly of pUC19. It is designed such that 5uL of the Positive Control DNA Mix is to be added to 15uL of Gibson Assembly Master Mix along side experimental reactions. Both pUC19 segments are between 1.3kb and 1.4kb in size. To construct the positive control reaction mix: | |||||||
| Changed: | ||||||||
| < < | AmpR half of pUC19, ~1150bp
| |||||||
| > > |
| |||||||
| Added: | ||||||||
| > > |
| |||||||
| Changed: | ||||||||
| < < | Ori half of pUC19, ~1600bp | |||||||
| > > | Using 5uL of this reaction provides approximately 1ng/100bp of each fragment in the Gibson Assembly reaction. | |||||||
| Deleted: | ||||||||
| < < |
| |||||||
| Changed: | ||||||||
| < < | The following PCR program works if using phusion polymerase, and will work for synthesizing both halves. | |||||||
| > > | -- Main.BrianRenda - 23 May 2014 | |||||||
| Deleted: | ||||||||
| < < | Initial denaturation: 98°C for 30 seconds 25 Cycles of: 98°C for 10s, 57°C for 20s, 72°C for 60s final extension: 72°C for 5 minutes -- Main.NeilGottel - 13 Feb 2013 | |||||||
Revision 82013-03-07 - NeilGottel
Revision 72013-02-26 - NeilGottel
Gibson Assembly-Work In Progress, do not use yetBackground and DesignGibson Cloning is a technique of DNA construct assembly that allows one to join multiple linear segments into either one large linear segment or, if the segments contain the appropriate components and overlaps, an intact plasmid. This protocol follows the one-step ISO assembly of overlapping dsDNA protocol. In order to assemble segments of DNA via Gibson Cloning, they must contain at least 40bp of homology to the segment they are being joined to. For example, if one was to make a construct that was Seg1-Seg2-Seg3 from individual PCRs of Seg1, Seg2, and Seg3, the 3' end of Seg1 would need 40bp of homology to the 5' end of Seg2 and Seg3 would require it 5' end to have 40bp of homology to Seg2. For constructing a plasmid, use a linearized vector backbone as one of your segments.SuppliesIf mastermix aliquots are available, and less than 1 year old:
Primer Design Using GibthonProtocol
Expected ResultsThe positive control should yield ampicillin resistant colonies when transformed into E. coli.Common Problems/TroubleshootingPositive control detailsAmpR half of pUC19, ~1150bp
|
Revision 62013-02-19 - NeilGottel
Gibson Assembly-Work In Progress, do not use yetBackground and DesignGibson Cloning is a technique of DNA construct assembly that allows one to join multiple linear segments into either one large linear segment or, if the segments contain the appropriate components and overlaps, an intact plasmid. This protocol follows the one-step ISO assembly of overlapping dsDNA protocol. In order to assemble segments of DNA via Gibson Cloning, they must contain at least 40bp of homology to the segment they are being joined to. For example, if one was to make a construct that was Seg1-Seg2-Seg3 from individual PCRs of Seg1, Seg2, and Seg3, the 3' end of Seg1 would need 40bp of homology to the 5' end of Seg2 and Seg3 would require it 5' end to have 40bp of homology to Seg2. For constructing a plasmid, use a linearized vector backbone as one of your segments.SuppliesIf mastermix aliquots are available, and less than 1 year old:
Primer Design Using GibthonProtocol
Expected Results | ||||||||
| Changed: | ||||||||
| < < | The positive control should yield ampicillin resistant colonies when transformed into E. coli. If ran a 1% agarose gel, expected size is | |||||||
| > > | The positive control should yield ampicillin resistant colonies when transformed into E. coli. | |||||||
Common Problems/TroubleshootingPositive control detailsAmpR half of pUC19, ~1150bp
| ||||||||
Revision 52013-02-13 - NeilGottel
| ||||||||
| Changed: | ||||||||
| < < | Gibson Assembly-Work In Progress (Neil) | |||||||
| > > | Gibson Assembly-Work In Progress, do not use yet | |||||||
Background and DesignGibson Cloning is a technique of DNA construct assembly that allows one to join multiple linear segments into either one large linear segment or, if the segments contain the appropriate components and overlaps, an intact plasmid. This protocol follows the one-step ISO assembly of overlapping dsDNA protocol. In order to assemble segments of DNA via Gibson Cloning, they must contain at least 40bp of homology to the segment they are being joined to. For example, if one was to make a construct that was Seg1-Seg2-Seg3 from individual PCRs of Seg1, Seg2, and Seg3, the 3' end of Seg1 would need 40bp of homology to the 5' end of Seg2 and Seg3 would require it 5' end to have 40bp of homology to Seg2. For constructing a plasmid, use a linearized vector backbone as one of your segments.Supplies | ||||||||
| Changed: | ||||||||
| < < | If mastermix aliquots are available, and less than 2 years old: | |||||||
| > > | If mastermix aliquots are available, and less than 1 year old: | |||||||
| ||||||||
| Added: | ||||||||
| > > |
Primer Design Using Gibthon | |||||||
Protocol
Expected ResultsThe positive control should yield ampicillin resistant colonies when transformed into E. coli. If ran a 1% agarose gel, expected size isCommon Problems/TroubleshootingPositive control details | ||||||||
| Changed: | ||||||||
| < < | AmpR half of pUC19, bp | |||||||
| > > | AmpR half of pUC19, ~1150bp | |||||||
| ||||||||
| Changed: | ||||||||
| < < | Ori half of pUC19, bp | |||||||
| > > | Ori half of pUC19, ~1600bp | |||||||
| ||||||||
| Changed: | ||||||||
| < < | 25 Cycles of: 98°C for 10s, 60°C for 20s, 72°C for 60s | |||||||
| > > | 25 Cycles of: 98°C for 10s, (60°C for 20s, 72°C for 60s | |||||||
| final extension: 72°C for 5 minutes | ||||||||
| Changed: | ||||||||
| < < | -- Main.BrianRenda - 26 Sep 2011 -- Main.NeilGottel - 08 Feb 2013 | |||||||
| > > | -- Main.NeilGottel - 13 Feb 2013 | |||||||
Revision 42013-02-09 - NeilGottel
| ||||||||
| Changed: | ||||||||
| < < | Gibson Cloning-Work In Progress (Neil) | |||||||
| > > | Gibson Assembly-Work In Progress (Neil) | |||||||
Background and DesignGibson Cloning is a technique of DNA construct assembly that allows one to join multiple linear segments into either one large linear segment or, if the segments contain the appropriate components and overlaps, an intact plasmid. This protocol follows the one-step ISO assembly of overlapping dsDNA protocol. In order to assemble segments of DNA via Gibson Cloning, they must contain at least 40bp of homology to the segment they are being joined to. For example, if one was to make a construct that was Seg1-Seg2-Seg3 from individual PCRs of Seg1, Seg2, and Seg3, the 3' end of Seg1 would need 40bp of homology to the 5' end of Seg2 and Seg3 would require it 5' end to have 40bp of homology to Seg2. For constructing a plasmid, use a linearized vector backbone as one of your segments.SuppliesIf mastermix aliquots are available, and less than 2 years old:
Protocol | ||||||||
| Deleted: | ||||||||
| < < |
| |||||||
Expected Results | ||||||||
| Added: | ||||||||
| > > | The positive control should yield ampicillin resistant colonies when transformed into E. coli. If ran a 1% agarose gel, expected size is | |||||||
Common Problems/TroubleshootingPositive control details | ||||||||
| Changed: | ||||||||
| < < | AmpR half of pUC19: | |||||||
| > > | AmpR half of pUC19, bp | |||||||
| ||||||||
| Changed: | ||||||||
| < < | Ori half of pUC19: | |||||||
| > > | Ori half of pUC19, bp | |||||||
| ||||||||
| Added: | ||||||||
| > > | ||||||||
| -- Main.BrianRenda - 26 Sep 2011 | ||||||||
| Changed: | ||||||||
| < < | -- Main.NeilGottel - 10 Jan 2013 | |||||||
| > > | -- Main.NeilGottel - 08 Feb 2013 | |||||||
Revision 32013-02-08 - NeilGottel
Gibson Cloning-Work In Progress (Neil)Background and DesignGibson Cloning is a technique of DNA construct assembly that allows one to join multiple linear segments into either one large linear segment or, if the segments contain the appropriate components and overlaps, an intact plasmid. This protocol follows the one-step ISO assembly of overlapping dsDNA protocol. In order to assemble segments of DNA via Gibson Cloning, they must contain at least 40bp of homology to the segment they are being joined to. For example, if one was to make a construct that was Seg1-Seg2-Seg3 from individual PCRs of Seg1, Seg2, and Seg3, the 3' end of Seg1 would need 40bp of homology to the 5' end of Seg2 and Seg3 would require it 5' end to have 40bp of homology to Seg2. For constructing a plasmid, use a linearized vector backbone as one of your segments.Supplies | ||||||||
| Changed: | ||||||||
| < < | ||||||||
| > > | If mastermix aliquots are available, and less than 2 years old: | |||||||
| Changed: | ||||||||
| < < | Protocol | |||||||
| > > |
| |||||||
| Added: | ||||||||
| > > |
| |||||||
| Added: | ||||||||
| > > | If no mastermix aliquots remain, but there is still <1 year old isothermal (ISO) reaction buffer available: | |||||||
| Deleted: | ||||||||
| < < | This protocol assumes the presence of Gibson Master Mix. | |||||||
| Changed: | ||||||||
| < < |
| |||||||
| > > | ||||||||
| Added: | ||||||||
| > > |
If no mastermix aliquots remain, and there is no ISO buffer, or the current ISO buffer is more than a year old:
Protocol | |||||||
Expected ResultsCommon Problems/Troubleshooting | ||||||||
| Added: | ||||||||
| > > |
Positive control detailsAmpR half of pUC19:
| |||||||
| -- Main.BrianRenda - 26 Sep 2011 -- Main.NeilGottel - 10 Jan 2013 | ||||||||
Revision 22013-01-10 - NeilGottel
| ||||||||
| Added: | ||||||||
| > > | Gibson Cloning-Work In Progress (Neil) | |||||||
| Deleted: | ||||||||
| < < | Gibson Cloning | |||||||
Background and DesignGibson Cloning is a technique of DNA construct assembly that allows one to join multiple linear segments into either one large linear segment or, if the segments contain the appropriate components and overlaps, an intact plasmid. This protocol follows the one-step ISO assembly of overlapping dsDNA protocol. In order to assemble segments of DNA via Gibson Cloning, they must contain at least 40bp of homology to the segment they are being joined to. For example, if one was to make a construct that was Seg1-Seg2-Seg3 from individual PCRs of Seg1, Seg2, and Seg3, the 3' end of Seg1 would need 40bp of homology to the 5' end of Seg2 and Seg3 would require it 5' end to have 40bp of homology to Seg2. For constructing a plasmid, use a linearized vector backbone as one of your segments. | ||||||||
| Added: | ||||||||
| > > | Supplies | |||||||
ProtocolThis protocol assumes the presence of Gibson Master Mix.
| ||||||||
| Added: | ||||||||
| > > | Expected Results | |||||||
| Added: | ||||||||
| > > | Common Problems/Troubleshooting | |||||||
| -- Main.BrianRenda - 26 Sep 2011 | ||||||||
| Added: | ||||||||
| > > | -- Main.NeilGottel - 10 Jan 2013 | |||||||
Revision 12011-09-26 - BrianRenda
Gibson CloningBackground and DesignGibson Cloning is a technique of DNA construct assembly that allows one to join multiple linear segments into either one large linear segment or, if the segments contain the appropriate components and overlaps, an intact plasmid. This protocol follows the one-step ISO assembly of overlapping dsDNA protocol. In order to assemble segments of DNA via Gibson Cloning, they must contain at least 40bp of homology to the segment they are being joined to. For example, if one was to make a construct that was Seg1-Seg2-Seg3 from individual PCRs of Seg1, Seg2, and Seg3, the 3' end of Seg1 would need 40bp of homology to the 5' end of Seg2 and Seg3 would require it 5' end to have 40bp of homology to Seg2. For constructing a plasmid, use a linearized vector backbone as one of your segments.ProtocolThis protocol assumes the presence of Gibson Master Mix.
|
View topic | History: r16 < r15 < r14 < r13 | More topic actions...