
Difference: ProtocolsBTKMakeANewPartPlasmid (1 vs. 17)
Revision 172023-06-14 - JeffreyBarrick
Part Plasmid AssemblyOnce you have designed your part and either amplified with PCR or ordered the desired gBlock (as described here) you can proceed to the assembly step of the part plasmid itself. The example reaction below shows pYTK001 used as the entry vector for the reaction; however, this can be substituted for any other entry vector with requisite BsmBI cut sites. Once built, part plasmids can be assembled into transcriptional units. Protocol source: NEB (https://www.neb.com/protocols/2020/01/15/golden-gate-assembly-protocol-for-using-neb-golden-gate-assembly-kit-bsmbi-v2-neb-e1602)Assembly ReactionAccess the old non-kit golden gate assembly protocols here 1. Calculate the mass (in ng) required for 50 fmol of vector and 100 fmol of insert using NEB's NEBioCalculator: https://nebiocalculator.neb.com/#!/dsdnaamt 2. Set up the following reaction mix:
| ||||||||||||
| Changed: | ||||||||||||
| < < |
| |||||||||||
| > > |
| |||||||||||
| Deleted: | ||||||||||||
| < < |
| |||||||||||
Optionally, you may have high enough efficiency if inserting one PCR product into the entry vector with NEB's short protocol with no cycling instead of the longer protocol:
Expected Results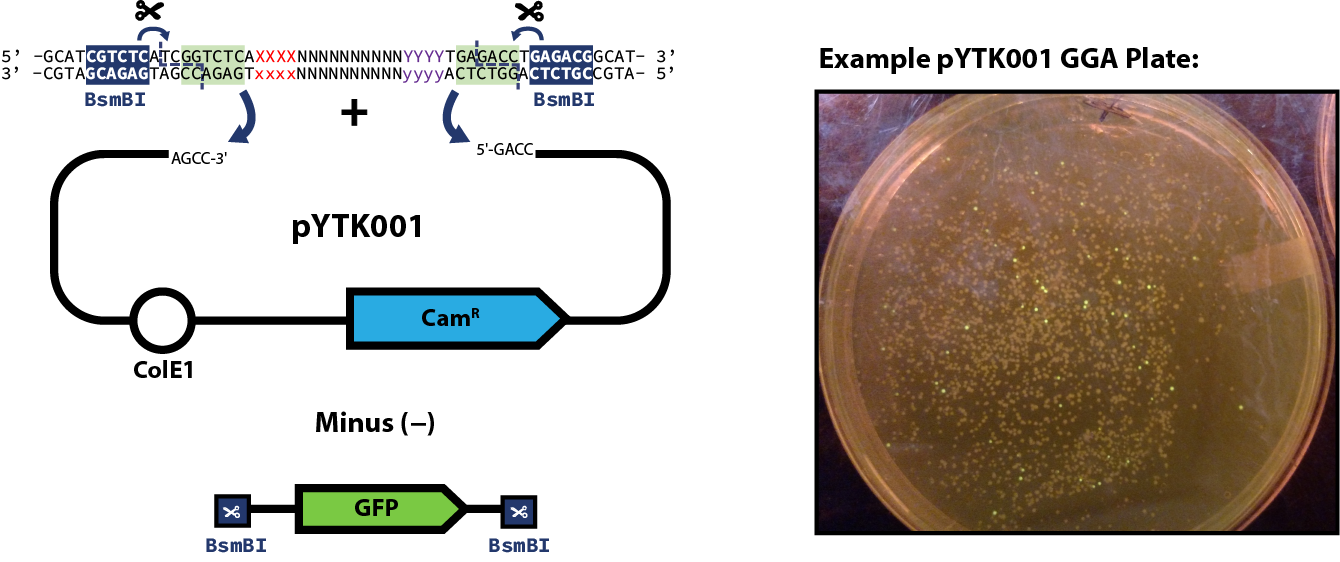
Back to Golden Gate Protocols
| ||||||||||||
Revision 162022-06-30 - JeffreyBarrick
Part Plasmid AssemblyOnce you have designed your part and either amplified with PCR or ordered the desired gBlock (as described here) you can proceed to the assembly step of the part plasmid itself. The example reaction below shows pYTK001 used as the entry vector for the reaction; however, this can be substituted for any other entry vector with requisite BsmBI cut sites. Once built, part plasmids can be assembled into transcriptional units. Protocol source: NEB (https://www.neb.com/protocols/2020/01/15/golden-gate-assembly-protocol-for-using-neb-golden-gate-assembly-kit-bsmbi-v2-neb-e1602)Assembly ReactionAccess the old non-kit golden gate assembly protocols here 1. Calculate the mass (in ng) required for 50 fmol of vector and 100 fmol of insert using NEB's NEBioCalculator: https://nebiocalculator.neb.com/#!/dsdnaamt 2. Set up the following reaction mix:
| |||||||||||||||||||||
| Added: | |||||||||||||||||||||
| > > |
Optionally, you may have high enough efficiency if inserting one PCR product into the entry vector with NEB's short protocol with no cycling instead of the longer protocol:
| ||||||||||||||||||||
4. Transform 2 μL assembly reaction into Electrocompetent or Chemically Competent cells and plate on LB + Cam
Expected Results
Back to Golden Gate Protocols
| |||||||||||||||||||||
Revision 152022-06-30 - JeffreyBarrick
Part Plasmid AssemblyOnce you have designed your part and either amplified with PCR or ordered the desired gBlock (as described here) you can proceed to the assembly step of the part plasmid itself. The example reaction below shows pYTK001 used as the entry vector for the reaction; however, this can be substituted for any other entry vector with requisite BsmBI cut sites. Once built, part plasmids can be assembled into transcriptional units. Protocol source: NEB (https://www.neb.com/protocols/2020/01/15/golden-gate-assembly-protocol-for-using-neb-golden-gate-assembly-kit-bsmbi-v2-neb-e1602)Assembly ReactionAccess the old non-kit golden gate assembly protocols here 1. Calculate the mass (in ng) required for 50 fmol of vector and 100 fmol of insert using NEB's NEBioCalculator: https://nebiocalculator.neb.com/#!/dsdnaamt 2. Set up the following reaction mix:
| |||||||||||||
| Changed: | |||||||||||||
| < < |
| ||||||||||||
| > > |
| ||||||||||||
| Added: | |||||||||||||
| > > |
| ||||||||||||
4. Transform 2 μL assembly reaction into Electrocompetent or Chemically Competent cells and plate on LB + Cam
Expected Results
Back to Golden Gate Protocols
| |||||||||||||
Revision 142021-11-03 - KateElston
Part Plasmid AssemblyOnce you have designed your part and either amplified with PCR or ordered the desired gBlock (as described here) you can proceed to the assembly step of the part plasmid itself. The example reaction below shows pYTK001 used as the entry vector for the reaction; however, this can be substituted for any other entry vector with requisite BsmBI cut sites. Once built, part plasmids can be assembled into transcriptional units. Protocol source: NEB (https://www.neb.com/protocols/2020/01/15/golden-gate-assembly-protocol-for-using-neb-golden-gate-assembly-kit-bsmbi-v2-neb-e1602)Assembly ReactionAccess the old non-kit golden gate assembly protocols here 1. Calculate the mass (in ng) required for 50 fmol of vector and 100 fmol of insert using NEB's NEBioCalculator: https://nebiocalculator.neb.com/#!/dsdnaamt 2. Set up the following reaction mix:
| ||||||||
| Changed: | ||||||||
| < < |
| |||||||
| > > |
| |||||||
Expected Results
Back to Golden Gate Protocols
| ||||||||
Revision 132021-11-03 - PatrickLariviere
Part Plasmid AssemblyOnce you have designed your part and either amplified with PCR or ordered the desired gBlock (as described here) you can proceed to the assembly step of the part plasmid itself. The example reaction below shows pYTK001 used as the entry vector for the reaction; however, this can be substituted for any other entry vector with requisite BsmBI cut sites. Once built, part plasmids can be assembled into transcriptional units. | |||||||||||||
| Added: | |||||||||||||
| > > | Protocol source: NEB (https://www.neb.com/protocols/2020/01/15/golden-gate-assembly-protocol-for-using-neb-golden-gate-assembly-kit-bsmbi-v2-neb-e1602) | ||||||||||||
Assembly ReactionAccess the old non-kit golden gate assembly protocols here | |||||||||||||
| Changed: | |||||||||||||
| < < | 1. Set up the following reaction mix: | ||||||||||||
| > > | 1. Calculate the mass (in ng) required for 50 fmol of vector and 100 fmol of insert using NEB's NEBioCalculator: https://nebiocalculator.neb.com/#!/dsdnaamt | ||||||||||||
| Added: | |||||||||||||
| > > | 2. Set up the following reaction mix: | ||||||||||||
| Changed: | |||||||||||||
| < < |
| ||||||||||||
| > > |
| ||||||||||||
| Deleted: | |||||||||||||
| < < |
| ||||||||||||
| |||||||||||||
| Changed: | |||||||||||||
| < < | 2. Mix samples well by pipetting, then run the reaction on the thermocycler under the following conditions:
| ||||||||||||
| > > | 3. Mix samples well by pipetting, then run the reaction on the thermocycler under the following conditions:
| ||||||||||||
| Deleted: | |||||||||||||
| < < |
| ||||||||||||
| Changed: | |||||||||||||
| < < | |||||||||||||
| > > | 4. Transform 2 μL assembly reaction into Electrocompetent or Chemically Competent cells and plate on LB + Cam | ||||||||||||
| Deleted: | |||||||||||||
| < < | 3. Transform 2 μL assembly reaction into Electrocompetent or Chemically Competent cells and plate on LB + Cam | ||||||||||||
Expected Results
Back to Golden Gate Protocols
| |||||||||||||
Revision 122021-11-03 - KateElston
Part Plasmid AssemblyOnce you have designed your part and either amplified with PCR or ordered the desired gBlock (as described here) you can proceed to the assembly step of the part plasmid itself. The example reaction below shows pYTK001 used as the entry vector for the reaction; however, this can be substituted for any other entry vector with requisite BsmBI cut sites. Once built, part plasmids can be assembled into transcriptional units.Assembly Reaction | |||||||||||||||||||||
| Added: | |||||||||||||||||||||
| > > | Access the old non-kit golden gate assembly protocols here | ||||||||||||||||||||
1. Set up the following reaction mix:
Expected Results
Back to Golden Gate Protocols
| |||||||||||||||||||||
Revision 112021-07-15 - KateElston
Part Plasmid AssemblyOnce you have designed your part and either amplified with PCR or ordered the desired gBlock (as described here) you can proceed to the assembly step of the part plasmid itself. The example reaction below shows pYTK001 used as the entry vector for the reaction; however, this can be substituted for any other entry vector with requisite BsmBI cut sites. Once built, part plasmids can be assembled into transcriptional units.Assembly Reaction1. Set up the following reaction mix:
Expected Results | |||||||||||||||||||||
| Added: | |||||||||||||||||||||
| > > |  | ||||||||||||||||||||
| Added: | |||||||||||||||||||||
| > > | |||||||||||||||||||||
Back to Golden Gate Protocols | |||||||||||||||||||||
| Added: | |||||||||||||||||||||
| > > |
| ||||||||||||||||||||
Revision 102021-07-09 - KateElston
Part Plasmid AssemblyOnce you have designed your part and either amplified with PCR or ordered the desired gBlock (as described here) you can proceed to the assembly step of the part plasmid itself. The example reaction below shows pYTK001 used as the entry vector for the reaction; however, this can be substituted for any other entry vector with requisite BsmBI cut sites. Once built, part plasmids can be assembled into transcriptional units. | |||||||||||||||||||
| Added: | |||||||||||||||||||
| > > | Assembly Reaction | ||||||||||||||||||
| Changed: | |||||||||||||||||||
| < < | Assembly reaction | ||||||||||||||||||
| > > | 1. Set up the following reaction mix: | ||||||||||||||||||
| Changed: | |||||||||||||||||||
| < < | Total volume will be 20 μL; You will need 10 fmol of entry vector and 20 fmol of your DNA insert(s). | ||||||||||||||||||
| > > | |||||||||||||||||||
| Deleted: | |||||||||||||||||||
| < < | |||||||||||||||||||
| |||||||||||||||||||
| Changed: | |||||||||||||||||||
| < < |
Mix samples well by pipetting, then run the reaction on the thermocycler under the following conditions: | ||||||||||||||||||
| > > |
| ||||||||||||||||||
| |||||||||||||||||||
| Added: | |||||||||||||||||||
| > > | |||||||||||||||||||
| Deleted: | |||||||||||||||||||
| < < |
| ||||||||||||||||||
| Added: | |||||||||||||||||||
| > > | 3. Transform 2 μL assembly reaction into Electrocompetent or Chemically Competent cells and plate on LB + Cam
Expected Results
| ||||||||||||||||||
Back to Golden Gate Protocols | |||||||||||||||||||
| Deleted: | |||||||||||||||||||
| < < | -- Main.KateElston - 29 Jan 2018 | ||||||||||||||||||
Revision 92021-06-29 - VictorLi
Part Plasmid AssemblyOnce you have designed your part and either amplified with PCR or ordered the desired gBlock (as described here) you can proceed to the assembly step of the part plasmid itself. The example reaction below shows pYTK001 used as the entry vector for the reaction; however, this can be substituted for any other entry vector with requisite BsmBI cut sites. Once built, part plasmids can be assembled into transcriptional units.Assembly reactionTotal volume will be 20 μL; You will need 10 fmol of entry vector and 20 fmol of your DNA insert(s).
Mix samples well by pipetting, then run the reaction on the thermocycler under the following conditions:
| |||||||||||||||||||||
| Added: | |||||||||||||||||||||
| > > |
| ||||||||||||||||||||
|
Back to Golden Gate Protocols -- Main.KateElston - 29 Jan 2018 | |||||||||||||||||||||
Revision 82021-06-17 - KateElston
| |||||||||||||||||||
| Changed: | |||||||||||||||||||
| < < | Back to Golden Gate Protocols | ||||||||||||||||||
| > > | Back to Golden Gate Protocols | ||||||||||||||||||
Part Plasmid AssemblyOnce you have designed your part and either amplified with PCR or ordered the desired gBlock (as described here) you can proceed to the assembly step of the part plasmid itself. The example reaction below shows pYTK001 used as the entry vector for the reaction; however, this can be substituted for any other entry vector with requisite BsmBI cut sites. Once built, part plasmids can be assembled into transcriptional units.Assembly reactionTotal volume will be 20 μL; You will need 10 fmol of entry vector and 20 fmol of your DNA insert(s).
Mix samples well by pipetting, then run the reaction on the thermocycler under the following conditions:
| |||||||||||||||||||
| Changed: | |||||||||||||||||||
| < < | Back to Golden Gate Protocols | ||||||||||||||||||
| > > | Back to Golden Gate Protocols | ||||||||||||||||||
-- Main.KateElston - 29 Jan 2018 | |||||||||||||||||||
Revision 72018-08-14 - KateElston
Part Plasmid AssemblyOnce you have designed your part and either amplified with PCR or ordered the desired gBlock (as described here) you can proceed to the assembly step of the part plasmid itself. The example reaction below shows pYTK001 used as the entry vector for the reaction; however, this can be substituted for any other entry vector with requisite BsmBI cut sites. Once built, part plasmids can be assembled into transcriptional units.Assembly reactionTotal volume will be 20 μL; You will need 10 fmol of entry vector and 20 fmol of your DNA insert(s). | |||||||||||||||||||
| Changed: | |||||||||||||||||||
| < < |
| ||||||||||||||||||
| > > |
| ||||||||||||||||||
Mix samples well by pipetting, then run the reaction on the thermocycler under the following conditions:
Back to Golden Gate Protocols -- Main.KateElston - 29 Jan 2018 | |||||||||||||||||||
Revision 62018-06-07 - DennisMishler
Part Plasmid AssemblyOnce you have designed your part and either amplified with PCR or ordered the desired gBlock (as described here) you can proceed to the assembly step of the part plasmid itself. The example reaction below shows pYTK001 used as the entry vector for the reaction; however, this can be substituted for any other entry vector with requisite BsmBI cut sites. Once built, part plasmids can be assembled into transcriptional units.Assembly reactionTotal volume will be 20 μL; You will need 10 fmol of entry vector and 20 fmol of your DNA insert(s).
Mix samples well by pipetting, then run the reaction on the thermocycler under the following conditions:
| |||||||||||||||||||||
| Added: | |||||||||||||||||||||
| > > | Back to Golden Gate Protocols | ||||||||||||||||||||
-- Main.KateElston - 29 Jan 2018 | |||||||||||||||||||||
Revision 52018-03-22 - KateElston
Part Plasmid AssemblyOnce you have designed your part and either amplified with PCR or ordered the desired gBlock (as described here) you can proceed to the assembly step of the part plasmid itself. The example reaction below shows pYTK001 used as the entry vector for the reaction; however, this can be substituted for any other entry vector with requisite BsmBI cut sites. Once built, part plasmids can be assembled into transcriptional units.Assembly reactionTotal volume will be 20 μL; You will need 10 fmol of entry vector and 20 fmol of your DNA insert(s).
| |||||||||||||||||||
| Changed: | |||||||||||||||||||
| < < | |||||||||||||||||||
| > > | |||||||||||||||||||
Mix samples well by pipetting, then run the reaction on the thermocycler under the following conditions:
-- Main.KateElston - 29 Jan 2018 | |||||||||||||||||||
Revision 42018-03-22 - SeanLeonard
Part Plasmid AssemblyOnce you have designed your part and either amplified with PCR or ordered the desired gBlock (as described here) you can proceed to the assembly step of the part plasmid itself. The example reaction below shows pYTK001 used as the entry vector for the reaction; however, this can be substituted for any other entry vector with requisite BsmBI cut sites. Once built, part plasmids can be assembled into transcriptional units.Assembly reactionTotal volume will be 20 μL; You will need 10 fmol of entry vector and 20 fmol of your DNA insert(s).
| ||||||||||
| Changed: | ||||||||||
| < < |
| |||||||||
| > > |
| |||||||||
| ||||||||||
| Changed: | ||||||||||
| < < | Mix samples well by pipetting, then run the reaction on the thermocycler under the following conditions: | |||||||||
| > > | Mix samples well by pipetting, then run the reaction on the thermocycler under the following conditions: | |||||||||
| ||||||||||
| Changed: | ||||||||||
| < < |
| |||||||||
| > > |
| |||||||||
| ||||||||||
| Changed: | ||||||||||
| < < |
| |||||||||
| > > |
| |||||||||
| Deleted: | ||||||||||
| < < | ||||||||||
| ||||||||||
| Changed: | ||||||||||
| < < | ||||||||||
| > > | ||||||||||
| Deleted: | ||||||||||
| < < | ||||||||||
| -- Main.KateElston - 29 Jan 2018 | ||||||||||
Revision 32018-01-30 - KateElston
| |||||||||||||||||||
| Added: | |||||||||||||||||||
| > > | Back to Golden Gate Protocols | ||||||||||||||||||
Part Plasmid AssemblyOnce you have designed your part and either amplified with PCR or ordered the desired gBlock (as described here) you can proceed to the assembly step of the part plasmid itself. The example reaction below shows pYTK001 used as the entry vector for the reaction; however, this can be substituted for any other entry vector with requisite BsmBI cut sites. Once built, part plasmids can be assembled into transcriptional units.Assembly reactionTotal volume will be 20 μL; You will need 10 fmol of entry vector and 20 fmol of your DNA insert(s).
| |||||||||||||||||||
| Changed: | |||||||||||||||||||
| < < | Back to Golden Gate Protocols | ||||||||||||||||||
| > > | |||||||||||||||||||
-- Main.KateElston - 29 Jan 2018 | |||||||||||||||||||
Revision 22018-01-29 - KateElston
Part Plasmid AssemblyOnce you have designed your part and either amplified with PCR or ordered the desired gBlock (as described here) you can proceed to the assembly step of the part plasmid itself. The example reaction below shows pYTK001 used as the entry vector for the reaction; however, this can be substituted for any other entry vector with requisite BsmBI cut sites. Once built, part plasmids can be assembled into transcriptional units.Assembly reactionTotal volume will be 20 μL; You will need 10 fmol of entry vector and 20 fmol of your DNA insert(s).
| |||||||||||||||||||||
| Changed: | |||||||||||||||||||||
| < < | |||||||||||||||||||||
| > > | Back to Golden Gate Protocols | ||||||||||||||||||||
-- Main.KateElston - 29 Jan 2018 | |||||||||||||||||||||
Revision 12018-01-29 - KateElston
Part Plasmid AssemblyOnce you have designed your part and either amplified with PCR or ordered the desired gBlock (as described here) you can proceed to the assembly step of the part plasmid itself. The example reaction below shows pYTK001 used as the entry vector for the reaction; however, this can be substituted for any other entry vector with requisite BsmBI cut sites. Once built, part plasmids can be assembled into transcriptional units.Assembly reactionTotal volume will be 20 μL; You will need 10 fmol of entry vector and 20 fmol of your DNA insert(s).
-- Main.KateElston - 29 Jan 2018 |
View topic | History: r17 < r16 < r15 < r14 | More topic actions...