
Difference: ProceduresStandardAgaroseGel (1 vs. 8)
Revision 82023-09-20 - IsaacGifford
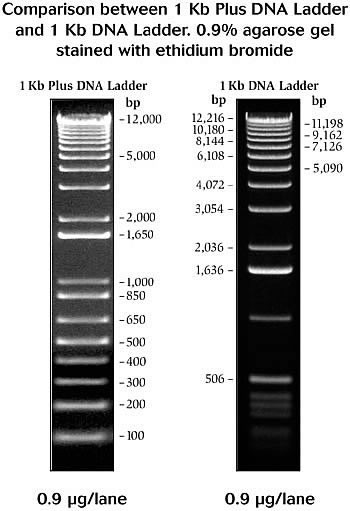
Gel ElectrophoresisOverviewGel electrophoresis separates DNA fragments based on size. Electric current moves the negatively charged DNA through the gel, which slows down larger pieces of DNA and allows smaller pieces to move faster. Bands on the gel can be compared to a "ladder" of DNA fragments of known lengths (Fig. 1) run in a separate well to determine the size of each piece of DNA.Fig. 1 NEB 1 kb and 1 kb+ ladders. Making the Gel | ||||||||
| Changed: | ||||||||
| < < | Most PCRs can be analyzed on a standard 1% agarose gel (1 g of agarose per 100 ml of buffer). Changing the concentration can be helpful for separating very large (use a 0.75% agarose gel) or very small (use a 2% agarose gel) DNA molecules. | |||||||
| > > | Most PCRs can be analyzed on a standard 1% agarose gel (1 g of agarose per 100 mL of buffer). Changing the concentration can be helpful for separating very large (use a 0.75% agarose gel) or very small (use a 2% agarose gel) DNA molecules. | |||||||
| For a standard 1% 50 ml gel: | ||||||||
| Changed: | ||||||||
| < < | 1. Add 0.5 g agarose and 50 ml TAE buffer to a 125 ml flask and heat in the microwave (on high power) for 1:30 minutes. While the microwave is running you can set up the gel mold and well combs. 2. After heating add 2.5 μl SYBR Safe and swirl to mix. | |||||||
| > > | 1. Add 0.5 g agarose and 50 mL TAE buffer to a 125 mL flask and heat in the microwave (on high power) for 1:30 minutes. While the microwave is running you can set up the gel mold and well combs. 2. After heating add 2.5 μL SYBR Safe and swirl to mix. | |||||||
| 3. Pour the liquid from the flask into the mold and let it solidify (approximately 20-30 minutes). | ||||||||
| Changed: | ||||||||
| < < | 4. Once the gel has solidified:
| |||||||
| > > | 4. Once the gel has solidified, remove the comb carefully so as not to break the walls of the wells. Then:
| |||||||
Loading and Running the GelDNA samples should be mixed with a loading dye before analyzing to help load them into the wells. Each sample and the ladder will then be pipetted into a separate well in the gel. Steps: 1. Pour TAE buffer into the rig until the gel is thoroughly submerged. | ||||||||
| Changed: | ||||||||
| < < | 2. Load 6 μl of 1 kb+ ladder working solution into the first well (dye is already combined with our ladder*). 3. Cut out a piece of parafilm to mix the loading dye and DNA samples. For each sample pipette a 1 μl drop of loading dye onto the parafilm. Then, mix 5 μl of PCR product into a drop of dye and pipette up and down to mix. Finally, pipette the dye/sample mix into an empty well. Record which lane each sample is loaded into. 4. Once all samples are loaded, attach the lid to the rig with the negative (black) electrode at the top and the positive (red) electrode at the bottom. (NOTE: always make sure that the current is off or paused before inserting or removing a cords from the power supply). 5. Set the voltage to 120 V and run the gel for about 20-30 minutes (you can set a timer on the machine itself by clicking on the clock logo. The run will stop when the time runs out). It is advisable to check up on the gel from time to time to make sure that it is proceeding normally. The tracking dye should migrate towards the bottom of the gel. | |||||||
| > > | 2. Load 6 μL of 1 kb+ ladder working solution into the first well (dye is already combined with our ladder*). 3. Cut out a piece of parafilm to mix the loading dye and DNA samples. For each sample pipette a 1 μL drop of loading dye onto the parafilm. Then, mix 5 μL of PCR product into a drop of dye and pipette up and down to mix. Finally, pipette the dye/sample mix into an empty well. Record which lane each sample is loaded into. 4. Once all samples are loaded, attach the lid to the rig with the negative (black) electrode at the top and the positive (red) electrode at the bottom. (NOTE: always make sure that the current is off or paused before inserting or removing cords from the power supply or rig). 5. Set the voltage to 120 V and run the gel for 20-30 minutes (you can set a timer on the machine itself by clicking on the clock logo. The run will stop when the time runs out). It is advisable to check up on the gel from time to time to make sure that it is proceeding normally. The tracking dye should migrate towards the bottom of the gel. | |||||||
6. When the gel seems to be completed, pause/stop the voltage, disconnect and remove the lid, and take the gel (in its tray) to the Bio-Rad gel imaging machine.
Note: Close PCR tubes when not being used. PCR products tend to dry up when exposed to air, leaving a lower volume that has a higher concentration of DNA.
*Our ladder working stock in the 4 °C fridge is colored purple from the loading dye and has been diluted as described here. Do not use the colorless undiluted stock, the bands will be too dense.
Imaging and Analyzing the GelSteps: 1. Remove the gel from the tray and place it in the imager. Open the Image Lab program. | ||||||||
| Changed: | ||||||||
| < < | 2. Select Protocol 1, then Position Gel, then OK on filter 2, then center your gel. 3. Once centered, close the imager door and select Run Protocol. | |||||||
| > > | 2. Select "SYBR Safe" from the "Nucleic Acids Gels" drop-down menu (unless using a different fluorescent dye), uncheck the "Highlight saturated pixels" box, then click "Position Gel." Center your gel in the viewer and zoom in to optimize your view of the gel. 3. Once centered, close the imager door and click "Run Protocol." | |||||||
| 4. Modify the image as you see fit**. 5. Save the image in Documents/[your name]. Note that the program saves files as .scn, which can only be opened by Image Lab and related programs. For a simpler version which can be viewed from any computer, select snapshot, then save a common file type (ex: .jpg). If you want a physical copy you can also print the image. **Image Lab contains several tools to aid in analyzing the gel image. Hitting the change contrast button allows manipulation of the brightness of the image, which may be helpful to clearly see faint bands. By clicking Lanes and Bands, then Automatic, the program puts labeled lanes where it perceives them. Bands can similarly be applied to the image. | ||||||||
| Changed: | ||||||||
| < < | Post-staining gels | |||||||
| > > | Post-staining Gels | |||||||
| Changed: | ||||||||
| < < | Adding DNA dyes to agarose gels after electrophoresis (“post-staining”) can be advantageous when very crisp bands are needed (publication, low amount of DNA, etc). Adding nucleic acid dye to the molten agarose can cause slight inhibit DNA migration and bending of bands. Post-staining can reduce bending, increase DNA signal, and reduce background signal | |||||||
| > > | Adding DNA dyes to agarose gels after electrophoresis (“post-staining”) can be advantageous when very crisp bands are needed (publication, low amount of DNA, etc). Adding nucleic acid dye to the molten agarose can slightly inhibit DNA migration and cause bands to bend. Post-staining can reduce bending, increase DNA signal, and reduce background signal. | |||||||
| Steps: 1. After gel electrophoresis, place gel into small tray. 2. Add 15 µL of dye into 50 mL buffer (typically x1 TAE) and add to tray. 3. Gently agitate for 30 minutes. 4. Remove buffer+dye. 5. Wash gel with ddH2O 2-3 times. 6. Image gel. *Notes:* 1. 15 µL of GelRed into 50 mL buffer is the correct dilution. | ||||||||
| Changed: | ||||||||
| < < | 2. SYBR-safe gel documentation notes that 50 µL of dye should be added to 50 mL buffer, though 15 µL of dye into 50 mL like likely sufficient (not tested). | |||||||
| > > | 2. SYBR-safe gel documentation notes that 50 µL of dye should be added to 50 mL buffer, though 15 µL of dye into 50 mL is likely sufficient (not tested). | |||||||
| 3. Typically, dye is diluted in the same buffer that the gel is made of/was run in (e.g.: x1 TAE). | ||||||||
| Changed: | ||||||||
| < < | 4. any volume of buffer+dye can be used, though it must completely cover the gel. | |||||||
| > > | 4. Any volume of buffer+dye can be used, though it must completely cover the gel. | |||||||
5. Post-staining buffer can be reused up to 5 times by storing in dark storage container (e.g.: a 50 mL conical tube covered with aluminum foil).
| ||||||||
Revision 72023-09-20 - IsaacGifford
Gel ElectrophoresisOverview | ||||||||
| Changed: | ||||||||
| < < | Gel electrophoresis separates DNA fragments based on size. Electric current moves the negatively charged DNA through the gel, which slows down larger pieces of DNA and allows smaller pieces to move faster. Bands on the gel can be compared to a "ladder" of DNA fragments of known lengths run in a separate well to determine the size of each piece of DNA. | |||||||
| > > | Gel electrophoresis separates DNA fragments based on size. Electric current moves the negatively charged DNA through the gel, which slows down larger pieces of DNA and allows smaller pieces to move faster. Bands on the gel can be compared to a "ladder" of DNA fragments of known lengths (Fig. 1) run in a separate well to determine the size of each piece of DNA. | |||||||
| Added: | ||||||||
| > > | Fig. 1 NEB 1 kb and 1 kb+ ladders. | |||||||
Making the Gel | ||||||||
| Changed: | ||||||||
| < < | Most PCRs can be analyzed on a standard 1% agarose gel (1g of agarose per 100 ml of buffer). Changing the concentration can be helpful for separating very large (use a 0.5% agarose gel) or very large size (use a 2% agarose gel). | |||||||
| > > | Most PCRs can be analyzed on a standard 1% agarose gel (1 g of agarose per 100 ml of buffer). Changing the concentration can be helpful for separating very large (use a 0.75% agarose gel) or very small (use a 2% agarose gel) DNA molecules. | |||||||
| For a standard 1% 50 ml gel: | ||||||||
| Changed: | ||||||||
| < < | 1. Add 0.5 g agarose and 50 ml TAE buffer to a 125 ml flask and heat in the microwave for 1:30 minutes. While the microwave is running you can set up the gel mold and well combs. | |||||||
| > > | 1. Add 0.5 g agarose and 50 ml TAE buffer to a 125 ml flask and heat in the microwave (on high power) for 1:30 minutes. While the microwave is running you can set up the gel mold and well combs. | |||||||
| 2. After heating add 2.5 μl SYBR Safe and swirl to mix. 3. Pour the liquid from the flask into the mold and let it solidify (approximately 20-30 minutes). 4. Once the gel has solidified: | ||||||||
| Changed: | ||||||||
| < < |
| |||||||
| > > |
| |||||||
| ||||||||
| Changed: | ||||||||
| < < | Loading the Gel | |||||||
| > > | Loading and Running the Gel | |||||||
| Changed: | ||||||||
| < < | DNA samples should be mixed with a loading dye before loading. | |||||||
| > > | DNA samples should be mixed with a loading dye before analyzing to help load them into the wells. Each sample and the ladder will then be pipetted into a separate well in the gel. | |||||||
| Added: | ||||||||
| > > | Steps: 1. Pour TAE buffer into the rig until the gel is thoroughly submerged. 2. Load 6 μl of 1 kb+ ladder working solution into the first well (dye is already combined with our ladder*). 3. Cut out a piece of parafilm to mix the loading dye and DNA samples. For each sample pipette a 1 μl drop of loading dye onto the parafilm. Then, mix 5 μl of PCR product into a drop of dye and pipette up and down to mix. Finally, pipette the dye/sample mix into an empty well. Record which lane each sample is loaded into. 4. Once all samples are loaded, attach the lid to the rig with the negative (black) electrode at the top and the positive (red) electrode at the bottom. (NOTE: always make sure that the current is off or paused before inserting or removing a cords from the power supply). 5. Set the voltage to 120 V and run the gel for about 20-30 minutes (you can set a timer on the machine itself by clicking on the clock logo. The run will stop when the time runs out). It is advisable to check up on the gel from time to time to make sure that it is proceeding normally. The tracking dye should migrate towards the bottom of the gel. 6. When the gel seems to be completed, pause/stop the voltage, disconnect and remove the lid, and take the gel (in its tray) to the Bio-Rad gel imaging machine. | |||||||
| Deleted: | ||||||||
| < < | First, gather all PCR products that are to be run, an appropriately sized ladder, and 6x loading dye. The latter two may be found in the 4 °C fridge in the computer room adjacent to the gel area. The 1 kb+ ladder is a combination 100 bp and 1 kb ladder which should be appropriate for any PCR sample (100 bp ladders are quite useful for samples less than 1500 bp, while 1 kb ladders are best suited for larger samples).
 Basic Steps:
1. Load 6 μl of 1 kb+ ladder into the first well (dye is already combined with ladder).
2. Combine dye and DNA on a cut out a sheet of parafilm. For each sample pipette 1 μl of loading dye onto the parafilm. Then, mix 5 μl of PCR product into a drop of dye and pipette up and down to mix. Finally, pipette the dye/sample mix into an empty well. Record which lane each sample is loaded into.
3. Once all samples are loaded, attach the electrodes to the rig with the negative (black) electrode at the top and the positive (red) electrode at the bottom. Start the run from the power unit.
Basic Steps:
1. Load 6 μl of 1 kb+ ladder into the first well (dye is already combined with ladder).
2. Combine dye and DNA on a cut out a sheet of parafilm. For each sample pipette 1 μl of loading dye onto the parafilm. Then, mix 5 μl of PCR product into a drop of dye and pipette up and down to mix. Finally, pipette the dye/sample mix into an empty well. Record which lane each sample is loaded into.
3. Once all samples are loaded, attach the electrodes to the rig with the negative (black) electrode at the top and the positive (red) electrode at the bottom. Start the run from the power unit. | |||||||
| Note: Close PCR tubes when not being used. PCR products tend to dry up when exposed to air, leaving a lower volume that has a higher concentration of DNA. | ||||||||
| Changed: | ||||||||
| < < | Running the Gel | |||||||
| > > | *Our ladder working stock in the 4 °C fridge is colored purple from the loading dye and has been diluted as described here. Do not use the colorless undiluted stock, the bands will be too dense. | |||||||
| Deleted: | ||||||||
| < < | Steps: 1. Once all samples have been loaded, attach a lid to the rig, and attach the lid to the Bio-Rad power supply (NOTE: always make sure that the current is off or paused before inserting or removing a cords from the power supply). 2. Set the voltage to 120 V and run the gel for about 20-30 minutes (you can set a timer on the machine itself by clicking on the clock logo. The run will stop when the time runs out). It is advisable to check up on the gel from time to time to make sure that it is proceeding normally. The tracking dye should migrate towards the bottom of the gel. 3. When the gel seems to be completed, pause/stop the voltage, disconnect and remove the lid, and take the gel (in its tray) to the Bio-Rad gel imaging machine. | |||||||
Imaging and Analyzing the GelSteps: | ||||||||
| Changed: | ||||||||
| < < | 1. Place the tray in the imager and open the Image lab program. | |||||||
| > > | 1. Remove the gel from the tray and place it in the imager. Open the Image Lab program. | |||||||
| 2. Select Protocol 1, then Position Gel, then OK on filter 2, then center your gel. 3. Once centered, close the imager door and select Run Protocol. | ||||||||
| Changed: | ||||||||
| < < | 4. Modify the image as you see fit*. | |||||||
| > > | 4. Modify the image as you see fit**. | |||||||
| 5. Save the image in Documents/[your name]. Note that the program saves files as .scn, which can only be opened by Image Lab and related programs. For a simpler version which can be viewed from any computer, select snapshot, then save a common file type (ex: .jpg). If you want a physical copy you can also print the image. | ||||||||
| Changed: | ||||||||
| < < | *Image lab contains several tools to aid in analyzing the gel image. Hitting the change contrast button allows manipulation of the brightness of the image, which may be helpful to clearly see faint bands. By clicking Lanes and Bands, then Automatic, the program puts labeled lanes where it perceives them. Bands can similarly be applied to the image. | |||||||
| > > | **Image Lab contains several tools to aid in analyzing the gel image. Hitting the change contrast button allows manipulation of the brightness of the image, which may be helpful to clearly see faint bands. By clicking Lanes and Bands, then Automatic, the program puts labeled lanes where it perceives them. Bands can similarly be applied to the image. | |||||||
| Changed: | ||||||||
| < < | Gel post-staining | |||||||
| > > | Post-staining gels | |||||||
| Adding DNA dyes to agarose gels after electrophoresis (“post-staining”) can be advantageous when very crisp bands are needed (publication, low amount of DNA, etc). Adding nucleic acid dye to the molten agarose can cause slight inhibit DNA migration and bending of bands. Post-staining can reduce bending, increase DNA signal, and reduce background signal Steps: 1. After gel electrophoresis, place gel into small tray. 2. Add 15 µL of dye into 50 mL buffer (typically x1 TAE) and add to tray. 3. Gently agitate for 30 minutes. 4. Remove buffer+dye. 5. Wash gel with ddH2O 2-3 times. 6. Image gel. *Notes:* | ||||||||
| Changed: | ||||||||
| < < | 1. 15 µL of GelRed into 50 mL buffer is the correct dilution 2. SYBR-safe gel documentation notes that 50 µL of dye should be added to 50 mL buffer, though 15 µL of dye into 50 mL like likely sufficient (not tested) 3. Typically, dye is diluted in the same buffer that the gel is made of/was run in (e.g.: x1 TAE) 4. any volume of buffer+dye can be used, though it must completely cover the gel 5. Post-staining buffer can be reused up to 5 times by storing in dark storage container (e.g.: a 50 mL conical tube covered with aluminum foil) | |||||||
| > > | 1. 15 µL of GelRed into 50 mL buffer is the correct dilution. 2. SYBR-safe gel documentation notes that 50 µL of dye should be added to 50 mL buffer, though 15 µL of dye into 50 mL like likely sufficient (not tested). 3. Typically, dye is diluted in the same buffer that the gel is made of/was run in (e.g.: x1 TAE). 4. any volume of buffer+dye can be used, though it must completely cover the gel. 5. Post-staining buffer can be reused up to 5 times by storing in dark storage container (e.g.: a 50 mL conical tube covered with aluminum foil). | |||||||
| ||||||||
Revision 62023-09-20 - IsaacGifford
Gel Electrophoresis | ||||||||
| Changed: | ||||||||
| < < | In order to analyze PCR results, the products are run on an agarose gel, which is then analyzed in UV light to ascertain the length of DNA fragments in the PCR reaction. | |||||||
| > > | Overview | |||||||
| Added: | ||||||||
| > > | Gel electrophoresis separates DNA fragments based on size. Electric current moves the negatively charged DNA through the gel, which slows down larger pieces of DNA and allows smaller pieces to move faster. Bands on the gel can be compared to a "ladder" of DNA fragments of known lengths run in a separate well to determine the size of each piece of DNA. | |||||||
Making the GelMost PCRs can be analyzed on a standard 1% agarose gel (1g of agarose per 100 ml of buffer). Changing the concentration can be helpful for separating very large (use a 0.5% agarose gel) or very large size (use a 2% agarose gel). | ||||||||
| Changed: | ||||||||
| < < | For a standard 1% 50 ml gel:
| |||||||
| > > | For a standard 1% 50 ml gel: 1. Add 0.5 g agarose and 50 ml TAE buffer to a 125 ml flask and heat in the microwave for 1:30 minutes. While the microwave is running you can set up the gel mold and well combs. 2. After heating add 2.5 μl SYBR Safe and swirl to mix. 3. Pour the liquid from the flask into the mold and let it solidify (approximately 20-30 minutes). 4. Once the gel has solidified: | |||||||
Loading the GelDNA samples should be mixed with a loading dye before loading. First, gather all PCR products that are to be run, an appropriately sized ladder, and 6x loading dye. The latter two may be found in the 4 °C fridge in the computer room adjacent to the gel area. The 1 kb+ ladder is a combination 100 bp and 1 kb ladder which should be appropriate for any PCR sample (100 bp ladders are quite useful for samples less than 1500 bp, while 1 kb ladders are best suited for larger samples). | ||||||||
| Changed: | ||||||||
| < < | Basic Steps:
| |||||||
| > > | Basic Steps: 1. Load 6 μl of 1 kb+ ladder into the first well (dye is already combined with ladder). 2. Combine dye and DNA on a cut out a sheet of parafilm. For each sample pipette 1 μl of loading dye onto the parafilm. Then, mix 5 μl of PCR product into a drop of dye and pipette up and down to mix. Finally, pipette the dye/sample mix into an empty well. Record which lane each sample is loaded into. 3. Once all samples are loaded, attach the electrodes to the rig with the negative (black) electrode at the top and the positive (red) electrode at the bottom. Start the run from the power unit. | |||||||
Note: Close PCR tubes when not being used. PCR products tend to dry up when exposed to air, leaving a lower volume that has a higher concentration of DNA.
Running the Gel | ||||||||
| Changed: | ||||||||
| < < | Steps:
| |||||||
| > > | Steps: 1. Once all samples have been loaded, attach a lid to the rig, and attach the lid to the Bio-Rad power supply (NOTE: always make sure that the current is off or paused before inserting or removing a cords from the power supply). 2. Set the voltage to 120 V and run the gel for about 20-30 minutes (you can set a timer on the machine itself by clicking on the clock logo. The run will stop when the time runs out). It is advisable to check up on the gel from time to time to make sure that it is proceeding normally. The tracking dye should migrate towards the bottom of the gel. 3. When the gel seems to be completed, pause/stop the voltage, disconnect and remove the lid, and take the gel (in its tray) to the Bio-Rad gel imaging machine. | |||||||
Imaging and Analyzing the Gel | ||||||||
| Changed: | ||||||||
| < < | Steps:
| |||||||
| > > | Steps: 1. Place the tray in the imager and open the Image lab program. 2. Select Protocol 1, then Position Gel, then OK on filter 2, then center your gel. 3. Once centered, close the imager door and select Run Protocol. 4. Modify the image as you see fit*. 5. Save the image in Documents/[your name]. Note that the program saves files as .scn, which can only be opened by Image Lab and related programs. For a simpler version which can be viewed from any computer, select snapshot, then save a common file type (ex: .jpg). If you want a physical copy you can also print the image. | |||||||
*Image lab contains several tools to aid in analyzing the gel image. Hitting the change contrast button allows manipulation of the brightness of the image, which may be helpful to clearly see faint bands. By clicking Lanes and Bands, then Automatic, the program puts labeled lanes where it perceives them. Bands can similarly be applied to the image.
Gel post-stainingAdding DNA dyes to agarose gels after electrophoresis (“post-staining”) can be advantageous when very crisp bands are needed (publication, low amount of DNA, etc). Adding nucleic acid dye to the molten agarose can cause slight inhibit DNA migration and bending of bands. Post-staining can reduce bending, increase DNA signal, and reduce background signal | ||||||||
| Changed: | ||||||||
| < < | Steps:
| |||||||
| > > | Steps: 1. After gel electrophoresis, place gel into small tray. 2. Add 15 µL of dye into 50 mL buffer (typically x1 TAE) and add to tray. 3. Gently agitate for 30 minutes. 4. Remove buffer+dye. 5. Wash gel with ddH2O 2-3 times. 6. Image gel. | |||||||
| Changed: | ||||||||
| < < | Notes: | |||||||
| > > | *Notes:* | |||||||
1. 15 µL of GelRed into 50 mL buffer is the correct dilution
2. SYBR-safe gel documentation notes that 50 µL of dye should be added to 50 mL buffer, though 15 µL of dye into 50 mL like likely sufficient (not tested)
3. Typically, dye is diluted in the same buffer that the gel is made of/was run in (e.g.: x1 TAE)
4. any volume of buffer+dye can be used, though it must completely cover the gel
5. Post-staining buffer can be reused up to 5 times by storing in dark storage container (e.g.: a 50 mL conical tube covered with aluminum foil)
| ||||||||
Revision 52023-09-20 - IsaacGifford
Gel ElectrophoresisIn order to analyze PCR results, the products are run on an agarose gel, which is then analyzed in UV light to ascertain the length of DNA fragments in the PCR reaction.Making the GelMost PCRs can be analyzed on a standard 1% agarose gel (1g of agarose per 100 ml of buffer). Changing the concentration can be helpful for separating very large (use a 0.5% agarose gel) or very large size (use a 2% agarose gel). For a standard 1% 50 ml gel: | ||||||||
| Changed: | ||||||||
| < < |
| |||||||
| > > |
| |||||||
| ||||||||
| Changed: | ||||||||
| < < |
| |||||||
| > > |
| |||||||
| Added: | ||||||||
| > > |
| |||||||
| Deleted: | ||||||||
| < < | Once a gel has been poured, it can be saved for later use if wrapped tightly in cellophane and stored in the 4°C fridge. | |||||||
Loading the GelDNA samples should be mixed with a loading dye before loading. | ||||||||
| Changed: | ||||||||
| < < | First, gather all PCR products that are to be run, an appropriately sized ladder, and 6x loading dye. The latter two may be found in the 4°C fridge in the computer room adjacent to the gel area. The 1kb+ ladder is a combination 100bp and 1kb ladder which should be appropriate for any PCR sample (100bp ladders are quite useful for samples less than 1500bp, while 1kb ladders are best suited for larger samples). | |||||||
| > > | First, gather all PCR products that are to be run, an appropriately sized ladder, and 6x loading dye. The latter two may be found in the 4 °C fridge in the computer room adjacent to the gel area. The 1 kb+ ladder is a combination 100 bp and 1 kb ladder which should be appropriate for any PCR sample (100 bp ladders are quite useful for samples less than 1500 bp, while 1 kb ladders are best suited for larger samples). | |||||||
 Basic Steps:
Basic Steps: | ||||||||
| Changed: | ||||||||
| < < |
| |||||||
| > > |
| |||||||
Running the GelSteps:
| ||||||||
| Changed: | ||||||||
| < < |
| |||||||
| > > |
| |||||||
Imaging and Analyzing the GelSteps:
Gel post-stainingAdding DNA dyes to agarose gels after electrophoresis (“post-staining”) can be advantageous when very crisp bands are needed (publication, low amount of DNA, etc). Adding nucleic acid dye to the molten agarose can cause slight inhibit DNA migration and bending of bands. Post-staining can reduce bending, increase DNA signal, and reduce background signal Steps:
| ||||||||
Revision 42023-09-19 - IsaacGifford
Gel ElectrophoresisIn order to analyze PCR results, the products are run on an agarose gel, which is then analyzed in UV light to ascertain the length of DNA fragments in the PCR reaction.Making the Gel | ||||||||
| Changed: | ||||||||
| < < | A standard 1% agarose gel uses 1g of agarose for every 100 ml of buffer. Percentages ranging from 0.5-2% agarose can be used (lower percentages are better for separating larger products and higher is better for smaller products but 1% should work for most samples) | |||||||
| > > | Most PCRs can be analyzed on a standard 1% agarose gel (1g of agarose per 100 ml of buffer). Changing the concentration can be helpful for separating very large (use a 0.5% agarose gel) or very large size (use a 2% agarose gel). | |||||||
| For a standard 1% 50 ml gel: | ||||||||
| Changed: | ||||||||
| < < |
| |||||||
| > > |
| |||||||
| Added: | ||||||||
| > > | Once a gel has been poured, it can be saved for later use if wrapped tightly in cellophane and stored in the 4°C fridge. | |||||||
Loading the Gel | ||||||||
| Changed: | ||||||||
| < < | DNA samples are loaded into the wells of an agarose gel using a p20 or p10 pipettor. First, gather all PCR products that are to be run, an appropriately sized ladder, and 6x loading dye. The latter two may be found in the 4°C fridge in the computer room adjacent to the gel area. The 1kb+ ladder is a combination 100bp and 1kb ladder which should be appropriate for any PCR sample (100bp ladders are quite useful for samples less than 1500bp, while 1kb ladders are best suited for larger samples). | |||||||
| > > | DNA samples should be mixed with a loading dye before loading. | |||||||
| Added: | ||||||||
| > > | First, gather all PCR products that are to be run, an appropriately sized ladder, and 6x loading dye. The latter two may be found in the 4°C fridge in the computer room adjacent to the gel area. The 1kb+ ladder is a combination 100bp and 1kb ladder which should be appropriate for any PCR sample (100bp ladders are quite useful for samples less than 1500bp, while 1kb ladders are best suited for larger samples). | |||||||
 Basic Steps:
Basic Steps: | ||||||||
| Changed: | ||||||||
| < < |
| |||||||
| > > |
| |||||||
| Changed: | ||||||||
| < < | Variation: Instead of combining all of the samples and then loading them, it is possible to load each sample as its combined with the dye. To do this, pipette up and down as normal, then push the pipettor a little bit past the first stop, enough to suck up all of the combined sample and dye. Now load this into a well, taking care to avoid shooting too much air into the well, as this may displace the sample. This method minimizes the amount of time samples spend exposed to the air, and thus prevents them from partially drying out on the parafilm. | |||||||
| > > | Note: Close PCR tubes when not being used. PCR products tend to dry up when exposed to air, leaving a lower volume that has a higher concentration of DNA. | |||||||
| Deleted: | ||||||||
| < < | Note: Close PCR tubes when not being used. PCR products tend to dry up when exposed to air, leaving a lower volume that has a higher concentration of DNA. | |||||||
Running the GelSteps: | ||||||||
| Changed: | ||||||||
| < < |
| |||||||
| > > |
| |||||||
| Deleted: | ||||||||
| < < |
| |||||||
Imaging and Analyzing the GelSteps:
Gel post-stainingAdding DNA dyes to agarose gels after electrophoresis (“post-staining”) can be advantageous when very crisp bands are needed (publication, low amount of DNA, etc). Adding nucleic acid dye to the molten agarose can cause slight inhibit DNA migration and bending of bands. Post-staining can reduce bending, increase DNA signal, and reduce background signal Steps:
| ||||||||
Revision 32023-02-22 - MattMcGuffie
Gel ElectrophoresisIn order to analyze PCR results, the products are run on an agarose gel, which is then analyzed in UV light to ascertain the length of DNA fragments in the PCR reaction.Making the GelA standard 1% agarose gel uses 1g of agarose for every 100 ml of buffer. Percentages ranging from 0.5-2% agarose can be used (lower percentages are better for separating larger products and higher is better for smaller products but 1% should work for most samples) For a standard 1% 50 ml gel:
Loading the GelDNA samples are loaded into the wells of an agarose gel using a p20 or p10 pipettor. First, gather all PCR products that are to be run, an appropriately sized ladder, and 6x loading dye. The latter two may be found in the 4°C fridge in the computer room adjacent to the gel area. The 1kb+ ladder is a combination 100bp and 1kb ladder which should be appropriate for any PCR sample (100bp ladders are quite useful for samples less than 1500bp, while 1kb ladders are best suited for larger samples). Basic Steps:
Basic Steps:
Running the GelSteps:
Imaging and Analyzing the GelSteps:
| ||||||||
| Added: | ||||||||
| > > | Gel post-staining | |||||||
| Changed: | ||||||||
| < < | -- Main.KateElston - 07 Jun 2018 | |||||||
| > > | Adding DNA dyes to agarose gels after electrophoresis (“post-staining”) can be advantageous when very crisp bands are needed (publication, low amount of DNA, etc). Adding nucleic acid dye to the molten agarose can cause slight inhibit DNA migration and bending of bands. Post-staining can reduce bending, increase DNA signal, and reduce background signal | |||||||
| Added: | ||||||||
| > > |
Steps:
| |||||||
| ||||||||
Revision 22018-06-07 - KateElston
Revision 12011-06-17 - AurkoDasgupta
Gel ElectrophoresisIn order to analyze PCR results, the products are run on an agarose gel, which is then analyzed in UV light to ascertain the length of DNA fragments in the PCR reaction.Making the GelA standard 1% agarose gel uses 1g of agarose for every 100 ml of buffer. A different percentage may be used, and gels with less than 1% agarose may be used to clearly distinguish products of very similar sizes. For a standard 50 ml gel, add .5g agarose and 50 ml SB buffer to a 125ml flask and heat for 1:30 minutes. Meanwhile, assemble a gel tray into its holder and find a comb with an appropriate number of wells, then place the comb into the rig. After heating add 2.5 μl SYBR Safe (5μl SYBR Safe for every 100mL gel) and swirl to mix. Pour the liquid from the flask into the rig and wait about 30 minutes for it to solidify. Once the gel has solidified, remove the comb, loosen the gel tray and place the gel into a rig. Pour SB buffer until the gel is thoroughly submerged.Loading the GelDNA samples are loaded into the wells of an agarose gel using a p20 or p10 pipettor. First, gather all PCR products that are to be run, an appropriately sized ladder, and 6x loading dye. The latter two may be found in the 4°C fridge in the computer room adjacent to the gel area. 100bp ladders are quite useful for samples less than 1500bp, while 1kb ladders are best suited for larger samples. First, load 6-7 μl of ladder into the first well. The easiest way to combine dye and DNA is to cut out a sheet of parafilm and make a drop of 1 μl of dye onto the parafilm for each sample to be run. Next, add 5 μl of PCR product to the dye and pipette up and down to homogenize. Once all samples are combined with dye, load them into the gel, making note of what sample goes into what lane. Variation: Instead of combining all of the samples and then loading them, it is possible to load each sample as its combined with the dye. To do this, pipette up and down as normal, then push the pipettor a little bit past the first stop, enough to suck up all of the combined sample and dye. Now load this into a well, taking care to avoid shooting too much air into the well, as this may displace the sample. This method minimizes the amount of time samples spend exposed to the air, and thus prevents them from partially drying out on the parafilm. Note: Close PCR tubes when not being used. PCR products tend to dry up when exposed to air, leaving a lower volume that has a higher concentration of DNA.Running the GelOnce all samples have been loaded, attach a lid to the rig, and attach the lid to the BioRad power supply (NOTE: always make sure that the current is off or paused before inserting or removing a cords from the power supply). Set the voltage to 150V and run the gel for about 30 minutes. It is advisable to check up on the gel from time to time to make sure that it is proceeding normally. When the gel seems to be completed, pause/stop the voltage, disconnect and remove the lid, and take the gel (in its tray) to the Bio-Rad gel imaging machine in the adjacent room.Imaging and Analyzing the GelPlace the tray in the imager and open the Image lab program. Select Protocol 1, then Position Gel, then OK on filter 2, then center your gel. Once centered, close the imager door and select Run Protocol. Once the imager finishes analyzing the image, save the image in Documents/[your name]. Note that the program saves files as .scn, which can only be opened by Image Lab and related programs. For a simpler version which can be viewed from any computer, select snapshot, then save a common file type (ex: .jpg). Emailing the images to oneself is a useful way to keep track of them. Image lab contains several tools to aid in analyzing the gel image. Hitting the change contrast button allows manipulation of the brightness of the image, which may be helpful to clearly see faint bands. By clicking Lanes and Bands, then Automatic, the program puts labeled lanes where it perceives them. Bands can similarly be applied to the image. When done modifying the image, print it out (saving it again / taking another snapshot might also be a good idea) and paste the picture in a lab notebook. -- Main.AurkoDasgupta - 17 Jun 2011 |
View topic | History: r8 < r7 < r6 < r5 | More topic actions...