Difference: PrimerDesignBenchling (13 vs. 14)
Revision 142025-03-20 - CameronRoots
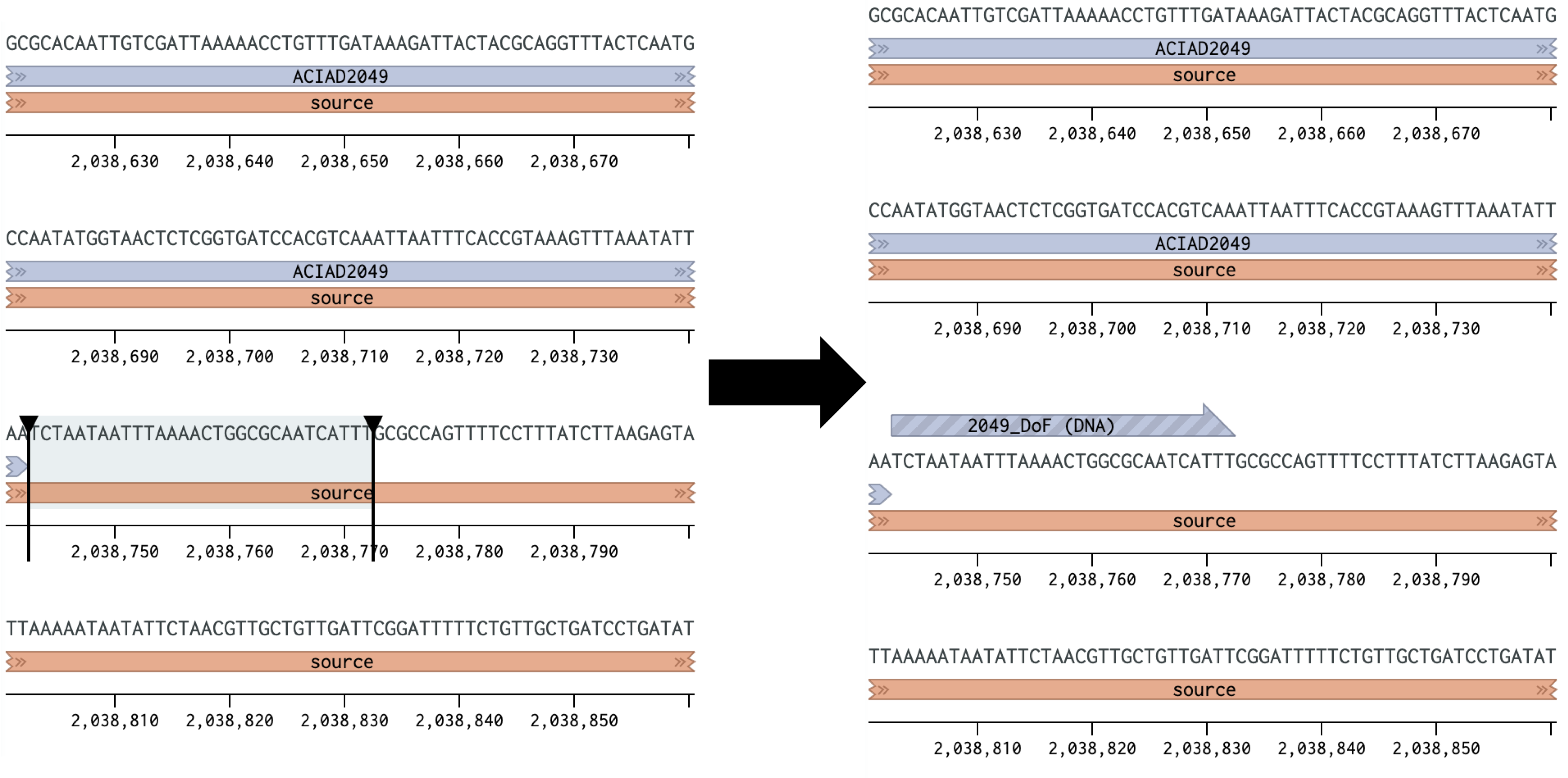
Custom Primer DesignOverviewWhile there are tools available for automatically designing primers (such as the NCBI Primer BLAST) often specialized PCR applications, such as amplifying fragments for Golden Gate or Gibson assembly or using different polymerases, will necessitate specific primer placements and modifications. In these cases it is important to take extra care to ensure the primers will anneal to the template and amplify well. This page presents guidelines and tools for specifically designing primers from DNA sequences, using the Benchling platform as an example. In short, you will need to: 1) Determine where to place your primers within the template sequence to amplify your desired product 2) Adjust your primer sequences to ideal annealing temperatures 3) Modify the sequences with any necessary adapters and check for inhibitory secondary structures Designing primers may require trade-offs and revisions to address specific issues that arise during the process. As such it's important to understand the needs of your specific experiment, such as specific locations in the genome, annealing temperatures, and adapter sequences, and keep those in mind while designing your primers. The examples below illustrate designing primers to amplify the downstream region of the Acinetobacter baylyi ADP1 ACIAD2049 gene for use in Golden Transformation.1) Determining Primer PlacementI. Start by identifying the region you are amplifying. Some common things to consider are:
Figure 1 Placing a forward primer downstream of the ACIAD2049 gene.
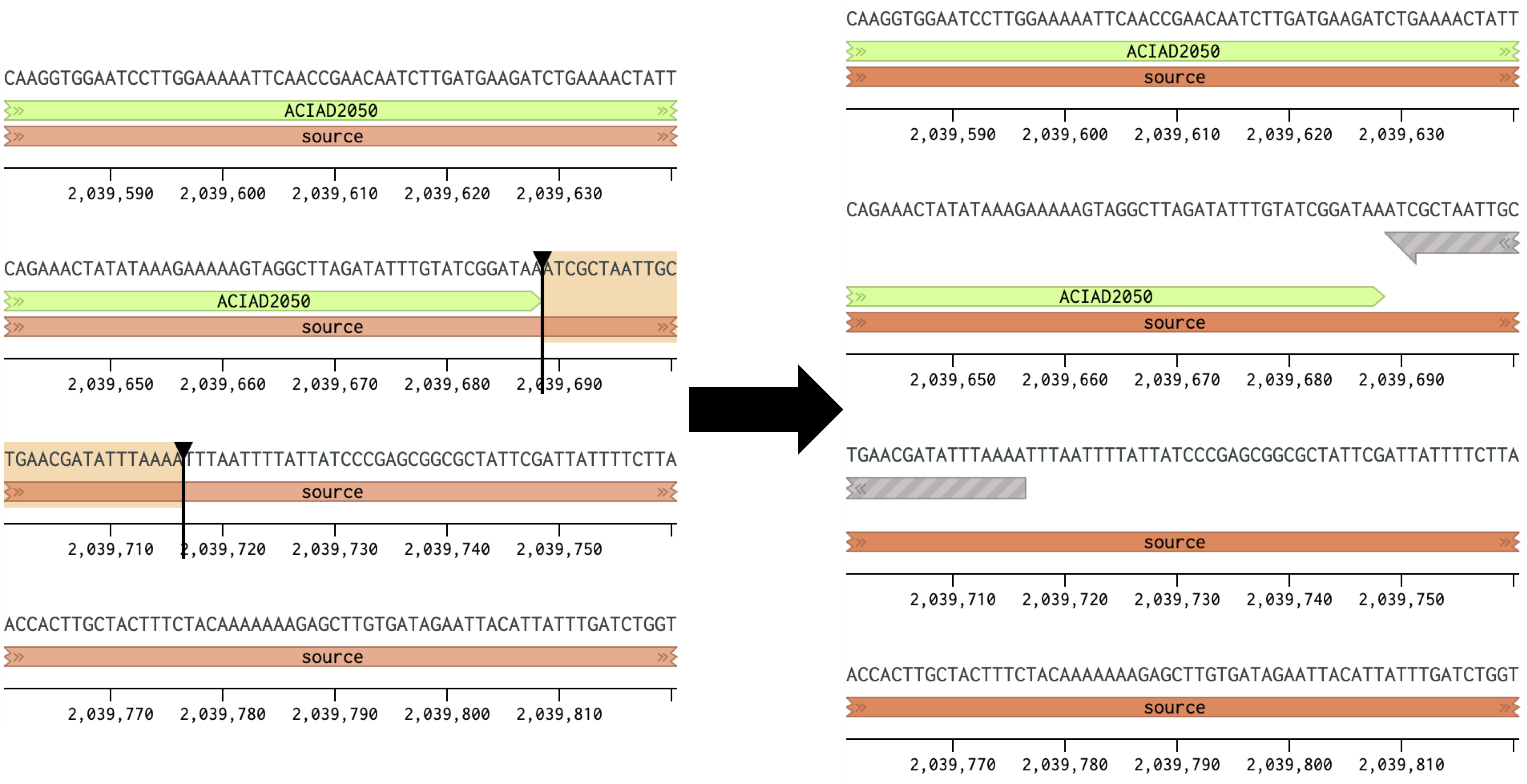
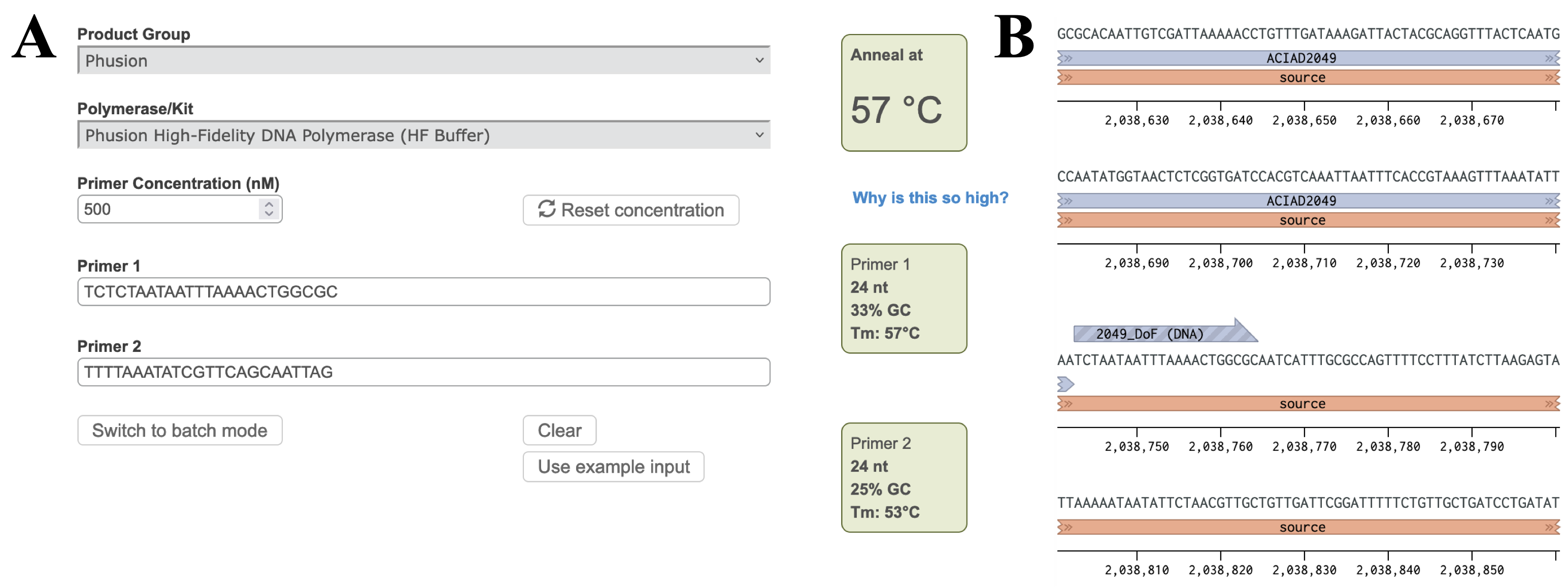
Figure 2 Placing a reverse primer downstream of the ACIAD2049 gene. In this example the primer is placed ~1000 bp downstream of the forward primer from Figure 1. 2) Calculating Annealing TemperaturesCalculate the annealing temperature for your PCR reaction using the NEB Tm Calculator. I. Select the polymerase for your PCR reaction from the drop-down menu. II. Copy ~30 bp of the forward and reverse primer sequences into the Primer 1 and Primer 2 boxes. III. Adjust the annealing temperature (Figure 3A, top right box) by removing bases from the 3' end of each primer until:
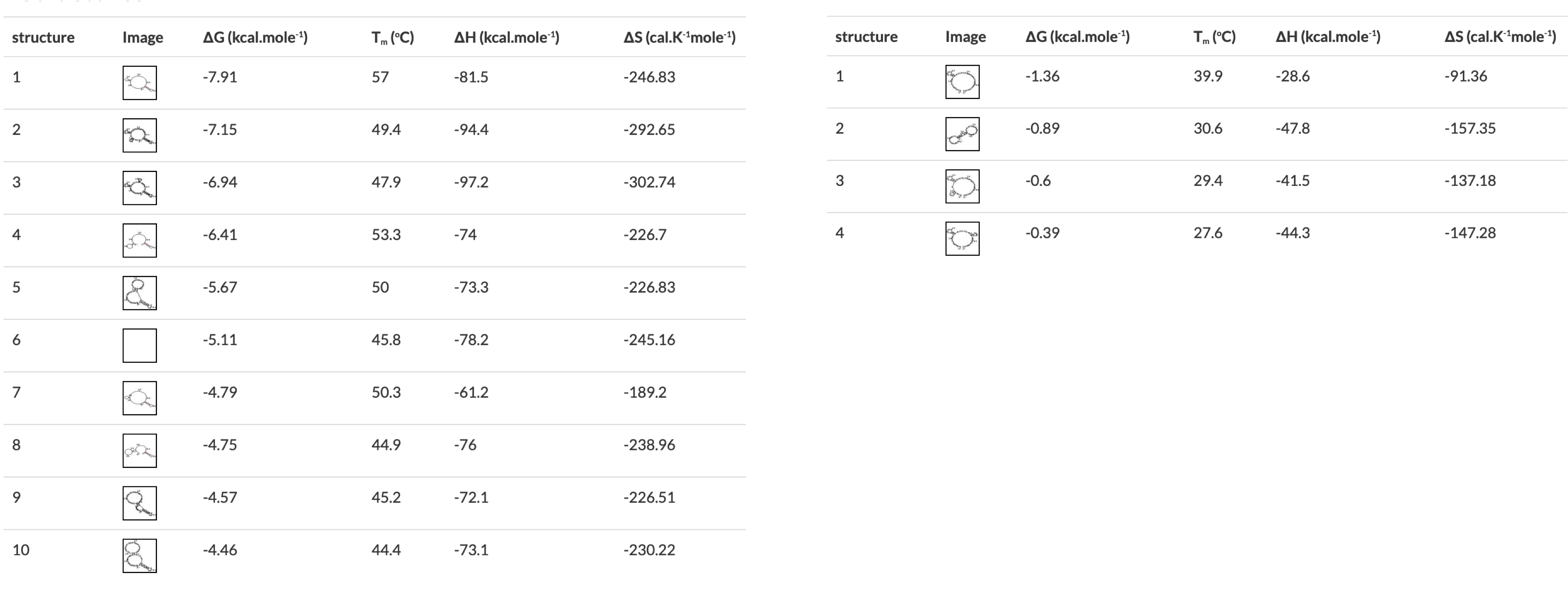
Figure 3 A) Annealing temperature calculations for ACIAD2049 downstream primers. B) ACIAD2049 downstream forward primer from Figure 1 shortened to account for annealing temperature. 3) Modifying Primer Sequences and Checking Secondary StructuresI. Add any additional modifications or adapters to the end of your sequences. Restriction sequences used for cloning need to be added to the 5' end of the primer sequence. II. Copy and paste each primer sequence into IDT's OligoAnalyzer program. Select "HAIRPIN" to analyze secondary structure formations. Structures with melting temperatures (Tm) below the annealing temperature for your reaction will melt in the thermocycler and not interfere with your reaction (Figure 4, left). Note any structures with Tms within a few degrees of your annealing temperature (Figure 4, right). III. If problematic structures were detected, modify the primer sequence to remove them. Potential solutions include:
Figure 4 OligoAnalyzer results showing primers with (left) and without (right) problematic secondary structures. The structures on the right would all melt in a typical PCR reaction, the first one on the left would form in a reaction with a 57░C annealing temperature. Figure 5 Checking for self-dimerization with OligoAnalyzer. In A the self-dimer is likely to interfere with PCR reactions but the self-dimer in B is not. C) Cartoon showing the extension of each primer by PCR (dotted line) resulting in a primer dimer from the primer in A. The self-dimer in B will not be extended by PCR. | |||||||||||||||||||||||||
| Added: | |||||||||||||||||||||||||
| > > |
4) Receipt and ResuspensionAs with most commercially sourced DNA, primers will typically arrive as a lyophilized film at the bottom of a cryo-tube. To use them, you must resuspend them in nuclease free H2O. Make a high-concentration stock by resuspending the lyophilized primer to a standard 100 ÁM concentration (that's micromolar = Ámol/L = pmol/Ál). The amount of primer in nanomoles (nmol or nm) is provided by the vendor on the spec sheet and the side of the tube. For example, for 28.5 nmol, add 285 Ál water to get a 100 ÁM stock solution. Pipette up and down until the film dissolves (you can watch it through the bottom of the tube) or resuspend by vortexing vigorously for 10 seconds. To prepare a 10 ÁM low-concentration working solution, take 10 Ál stock solution and add to 90 Ál water (or some multiple thereof, like 30 Ál of high concentration stock + 270 Ál water). If you choose to resuspend your primers to a concentration other than 100 ÁM, please write this clearly on the primer tube. If you are diluting from someone else's suspended primers, it wouldn't hurt to verify with them that the concentration is 100 ÁM before diluting. If you have a 200ÁM concentration, use 5ÁL primer in 95ÁL water to get a 10ÁM low-concentration working solution. Sometimes micromolar (Ámol/L) concentrations are abbreviated in brackets, e.g. [100] mean 100 micromolar. Store primers at -20░C for long term storage or -4░C for immediate use. | ||||||||||||||||||||||||
Further Reading
| |||||||||||||||||||||||||