
Difference: ResearchInterests (28 vs. 29)
Revision 292016-02-20 - JeffreyBarrick
<-- Preferences start here
Barrick Lab :: Research Active ProjectsPreventing Evolutionary Failure in Synthetic Biology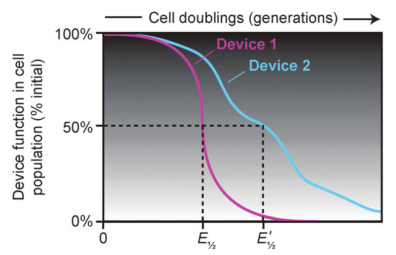 Evolutionary half-lives of biological devices
Dynamics of Microbial Genome Evolution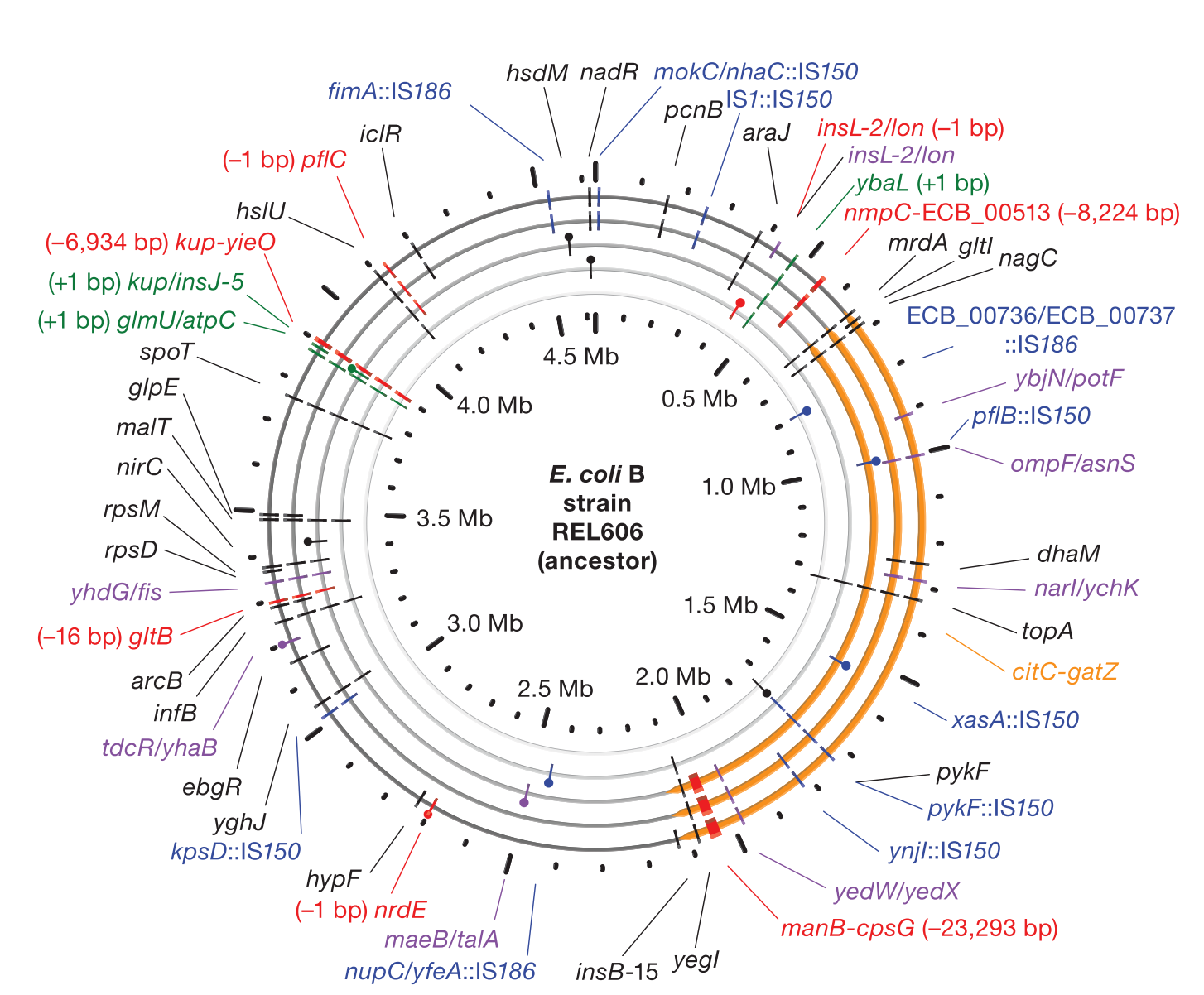 Accumulation of mutations in population Ara-1 of the LTEE over 20,000 generations of evolution | |||||||||||||||||||||
| Deleted: | |||||||||||||||||||||
| < < | Representative Publications
| ||||||||||||||||||||
| Resources | |||||||||||||||||||||
| Added: | |||||||||||||||||||||
| > > | Representative Publications
| ||||||||||||||||||||
Funding: NIH K99/R00, NSF, NSF BEACON Center, CPRIT
Identifying Mutations that Promote Bacterial Evolvability Epistasis Poker. Early draws of cards that are more beneficial to the value of a poker hand (analogous to the most beneficial mutations in the eventual loser E. coli), prevent them from reaching the best hand after more cards are drawn. 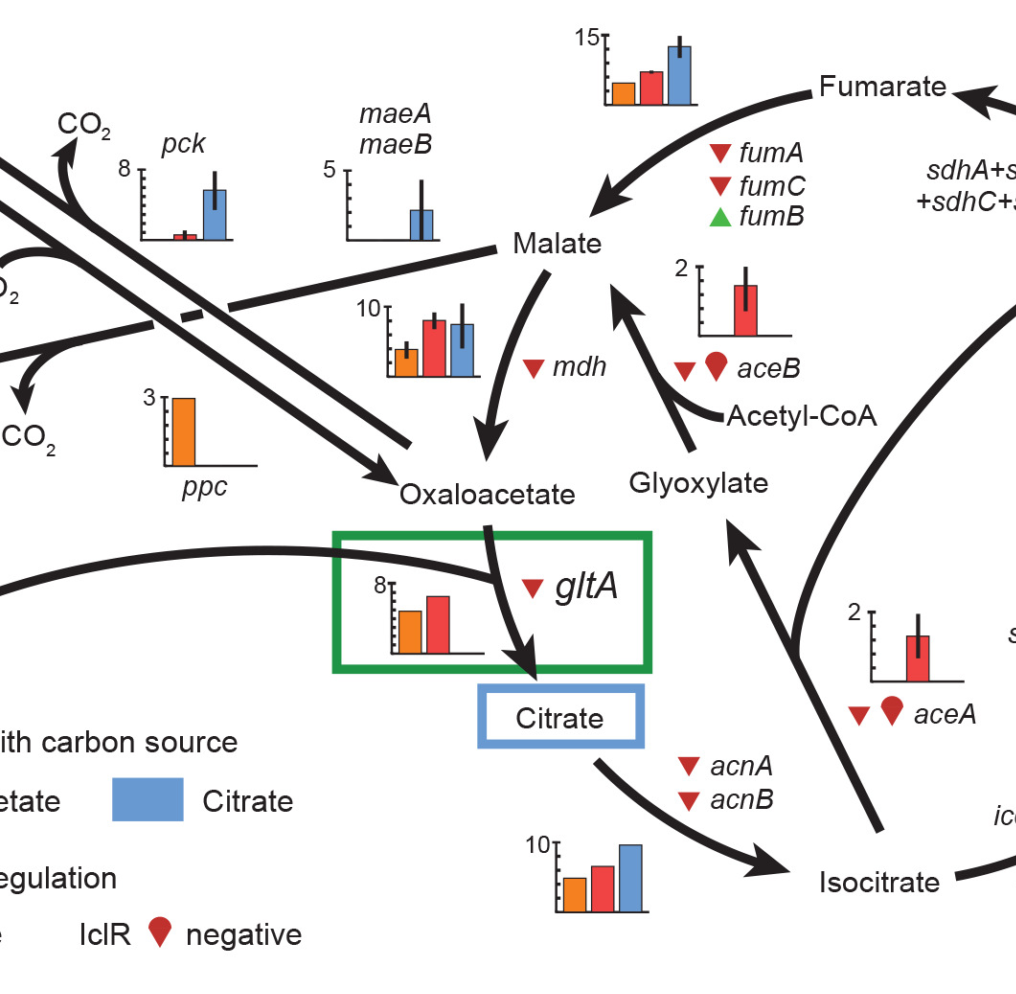 Evolution of central metabolism potentiates and refines a metabolic innovation
Evolving and Engineering Naturally Transformable BacteriaNaturally competent bacteria may have increased evolutionary potential because they can directly acquire new genes from their environments and incorporate them into their genomes. In addition to this possibility of a new mutational move in genotype-phenotype space (by horizontal gene transfer), these microbes provide an improved platform for studying microbial genome engineering and evolution due to the ease of reconstructing mutations and introducing new genes. We are investigating sources of genetic instability in the naturally competent bacterium Acinetobacter baylyi ADP1 and engineering a clean genome version of this strain to have reduced rates of mutations that lead to inactivation of introduced genes for synthetic biology applications. We are also using experimental evolution to test for the utility of providing foreign DNA sequences and to understand how horizontally acquired genes become domesticated after they are incorporated into a new genome. Finally, we are using ADP1 as a platform for understanding the limits of simplifying a bacterial genome by evolutionary streamlining approaches. Representative Publication
Experimental Evolution with Expanded Genetic CodesAnother way to potentially increase evolvability is to augment the chemical diversity of the building blocks available to an organism. We are evolving microorganisms, including bacteriophage, in the context of expanded genetic codes that re-code the amber stop codon into various non-canonical amino acids. This change increases the sequence-structure-function space available for evolution to explore. We are studying how evolution must compensate for the global change in how the information in an organism's genome is decoded, in cases where it is possible for an organism to survive this transition, and also how dependence on the new amino acid develops over time, such that an organism can no longer survive in the context of a normal genetic code. Finally, we are systematically examining how the unique chemistries of different amino acid building blocks are tolerated and uniquely useful for adaptation and using this technology to recapitulate the steps in a hypothesized pathway in which new post-translational modifications evolve in a series of adaptive steps from nonenzymatic metabolite damage to the proteome. Representative Publication
AG3C: Associating Growth Conditions with Cellular CompositionWe are part of a collaboration of eight labs (including experimental biologists, computational biologists, and mathematicians) that is collecting and analyzing comprehensive transcriptomics, proteomics, lipidomics, and metabolic flux data from bacterial samples grown under different conditions. The aim is to solve the "inverse problem" of predicting the conditions that were used to culture a microbial sample from measurements in these high-dimensional data sets (AG3C Website
Previous ProjectsDiscovering Functional Nucleic Acid Families by Deep Sequencing and Fold SamplingA final way that we are studying how to improve the potential of evolutionary methods is in systems for the in vitro selection of nucleic acid molecules. Nucleic acids with random sequences often misfold into inactive structures that are kinetically trapped. That is, they become stuck and cannot unfold to rearrange into other, potentially active, structures on a reasonable timescale. It is possible that this potential for misfolding, which is known to occur for even natural ribozymes, limits the recovery of new families of functional nucleic acids (aptamers, ribozymes, deoxyribozymes, riboswitches) by laboratory evolution methods that select for functional molecules from pools of 1013–1015 molecules with randomized sequences. We expect this problem to more greatly affect longer nucleic acid sequences, which have the capacity for folding into elaborate three-dimensional structures that achieve more rapid rates of catalysis or tighter binding affinities. We are currently using next-generation deep-sequencing techniques and methods to allow a single nucleic acid strand to have multiple chances to fold correctly to pass selection to see if they can enhance our ability to recover more complex families of functional nucleic acids. Representative Publications
Other InterestsEvolution Experiments with Digital OrganismsPopulations of self-replicating computer programs mutate and compete for CPU cycles in the Avida artificial life system. This environment for digital organisms has been used to ask fundamental questions that transcend the substrate that is evolving. The speed of digital evolution experiments, transparency of the underlying mechanics, and ability to manipulate and record every aspect of the evolutionary process make it possible to more readily explore cause and effect relationships in Avida than in natural systems. I have extensive experience with Avida. My lab will use Avida to complement studies in biological systems: both as a way of more clearly illustrating and exhaustively characterizing phenomena of interest and as a source of new candidate laws and insights to motivate new lines of experimental inquiry. ResourcesCollaboration InterestsIn addition to these specific plans for future research, we have long-standing interests in RNA biochemistry, riboswitches and other noncoding RNAs in bacteria, and comparative genomics. We hope to be a resource to colleagues who are interested in adding an evolutionary, informatics, or genomics dimension to their studies. Whenever possible, we would like to investigate our basic research questions about evolution in systems where there is additional interest in the underlying molecules, cells, and mutations. We are eager to link up with researchers and those in industry who have relevant problems in medicine and biotechnology to find these opportunities. Please contact Prof. Barrick about potential collaborations.
| |||||||||||||||||||||
View topic | History: r63 < r62 < r61 < r60 | More topic actions...
Ideas, requests, problems regarding TWiki? Send feedback