
Difference: ProtocolsGrowthRates (1 vs. 15)
Revision 152024-01-04 - JeffreyBarrick
Measuring Microbial Growth Rates in a Plate Reader | |||||||||
| Changed: | |||||||||
| < < | The following protocol can be used to determine the growth rate of a bacterial culture using a plate reader by measuring the optical density (OD600) of the culture over time. This protocol is written specifically for a Tecan Infinite® M200 Pro plate reader combined with the Magellan™ data analysis software and thus may require alteration for other machines. The Nunc™ MicroWell™ | ||||||||
| > > | The following protocol can be used to determine the growth rate of a bacterial culture using a plate reader by measuring the optical density (OD600) of the culture over time. This protocol is written specifically for a Tecan Infinite® M200 Pro plate reader combined with the Magellan™ data analysis software and thus may require alteration for other machines. The Nunc™ MicroWell™ 96-Well Optical-Bottom Plates (black wells with clear bottoms) are ideal for this assay. | ||||||||
| Deleted: | |||||||||
| < < | 96-Well Optical-Bottom Plates (black wells with clear bottoms) are ideal for this assay. | ||||||||
| Added: | |||||||||
| > > | Some considerations:
| ||||||||
Program set-upThe parameters used by the Barrick Lab are:
Reviving cultures (2 days before experiment)1) Inoculate an overnight culture of each strain being tested. Prepare a separate tube of uninoculated media as a control for contamination. Incubate overnight.Preconditioning cultures (1 day before experiment)Preconditioning acclimates the strains to the media. Additionally, on this day you should pre-warm your media as it takes a long time for the plate reader to warm up media. Not doing this will lead to inconsistent lag time. 2) Inoculate 5 μl of each overnight culture into fresh media to precondition. Prepare a separate tube of uninoculated media as a control for contamination. Incubate overnight.3) Place the media you will use for the assay in an incubator at the correct growth temperature overnight to pre-warm. Growing cells in plate reader and measuring OD600All strains tested should have at least have 3 replicates, although more replicates should be performed as long as there is available space on the plate. Evaporation can occur in the outermost wells, so if there are few enough samples the outer ring of wells should be skipped. Due to small variations in temperature throughout a 96-well plate, best results will be obtained if replicates are distributed randomly across the plate. 4) Begin heating the plate reader to the desired growth temperature using the “Temp. control...” button in the “Start Measurement” dialog box.
6) Inoculate each test well with 5 μl of overnight culture. 7) Place the plate into the plate reader. The lid can be removed (we've had no problems with contamination, as seen on wells with LB blanks, with this). 8) Start your program. 9) Once the program has finished, export the data as an Excel spreadsheet.
Formatting the dataIn order to run Growthcurver, you need to create a tab-delimited file in the following arrangement (see example file below):
Fitting growth curves with Growthcurver in RGrowthcurver is an R package that analyzes the optical density data by fitting it to a logistic function from which the growth rate, doubling time, and carrying capacity can be calculated. Growthcurver can be installed with the command:
install.packages("growthcurver")
Growthcurver can be run on the formatted tab-delimited file with the following code. The variable "r" contains the growth rate and is extracted once the model has been fit to the data. Note that the units of the growth rate will be based on the units of your file, i.e. if your time is in seconds and the growth measurements are in OD600 the growth rate will be expressed in OD600/second and may need to be converted to other units.
Quality control can be performed by checking for notes on the model fitting. Any samples with low quality fits will be identified by the following code:
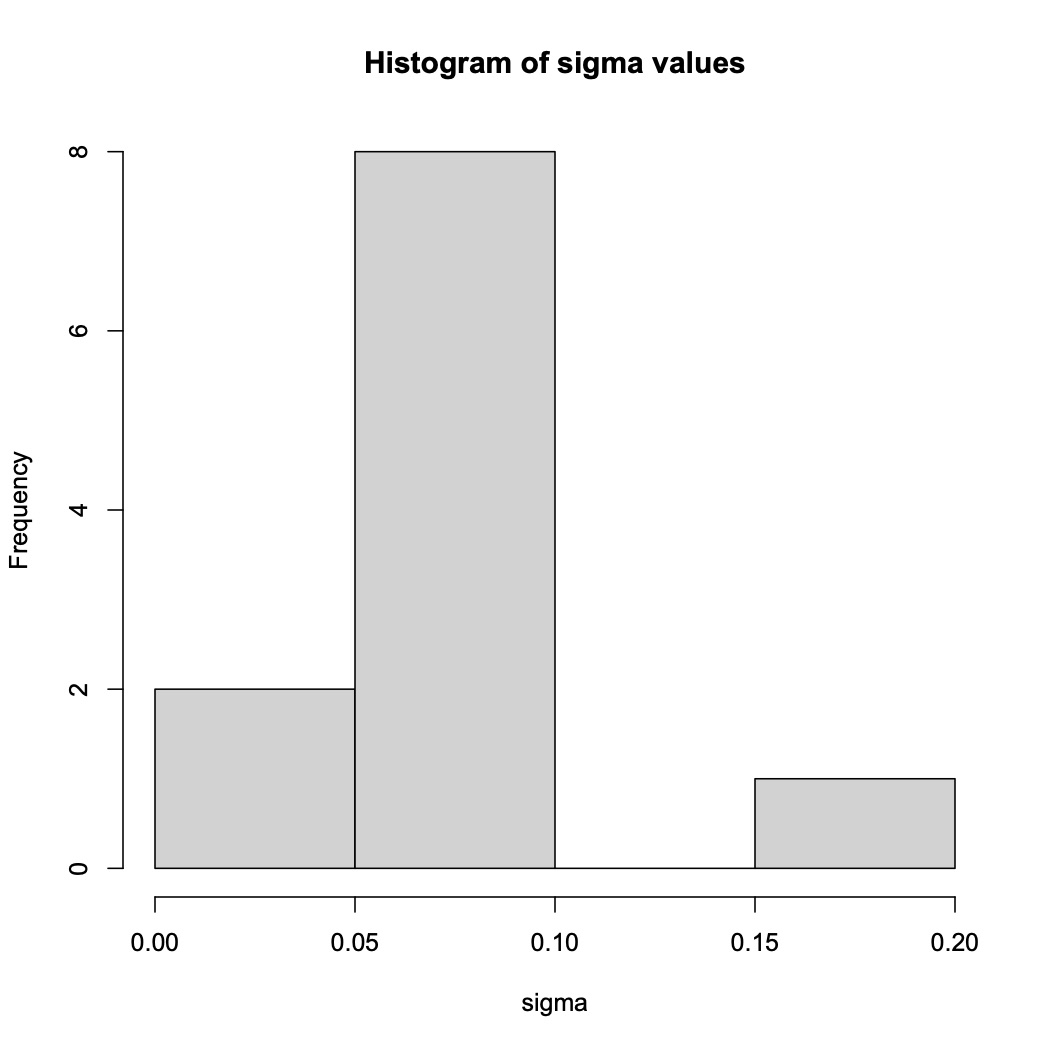
Other statistics that can be reported include the carrying capacity (use gc_out$k), the doubling time (gc_out$t_gen), and the inflection point of log phase (gc_out$t_mid) for each sample.Additionally, a histogram of the variance of the data can be produced with the following code. Poorly fit samples will appear as outliers on the histogram as in the example below.
Calculating growth rates using the Grofit R packageGrowth curves can also be fit using Grofit, however this package is no longer supported by the current version of R.References
| |||||||||
| Added: | |||||||||
| > > |
| ||||||||
Contributors
| |||||||||
Revision 142021-11-05 - IsaacGifford
Measuring Microbial Growth Rates in a Plate ReaderThe following protocol can be used to determine the growth rate of a bacterial culture using a plate reader by measuring the optical density (OD600) of the culture over time. This protocol is written specifically for a Tecan Infinite® M200 Pro plate reader combined with the Magellan™ data analysis software and thus may require alteration for other machines. The Nunc™ MicroWell™ 96-Well Optical-Bottom Plates (black wells with clear bottoms) are ideal for this assay.Program set-upThe parameters used by the Barrick Lab are:
Reviving cultures (2 days before experiment)1) Inoculate an overnight culture of each strain being tested. Prepare a separate tube of uninoculated media as a control for contamination. Incubate overnight.Preconditioning cultures (1 day before experiment)Preconditioning acclimates the strains to the media. Additionally, on this day you should pre-warm your media as it takes a long time for the plate reader to warm up media. Not doing this will lead to inconsistent lag time. 2) Inoculate 5 μl of each overnight culture into fresh media to precondition. Prepare a separate tube of uninoculated media as a control for contamination. Incubate overnight.3) Place the media you will use for the assay in an incubator at the correct growth temperature overnight to pre-warm. Growing cells in plate reader and measuring OD600All strains tested should have at least have 3 replicates, although more replicates should be performed as long as there is available space on the plate. Evaporation can occur in the outermost wells, so if there are few enough samples the outer ring of wells should be skipped. Due to small variations in temperature throughout a 96-well plate, best results will be obtained if replicates are distributed randomly across the plate. 4) Begin heating the plate reader to the desired growth temperature using the “Temp. control...” button in the “Start Measurement” dialog box.
6) Inoculate each test well with 5 μl of overnight culture. 7) Place the plate into the plate reader. The lid can be removed (we've had no problems with contamination, as seen on wells with LB blanks, with this). 8) Start your program. 9) Once the program has finished, export the data as an Excel spreadsheet.
Formatting the dataIn order to run Growthcurver, you need to create a tab-delimited file in the following arrangement (see example file below):
Fitting growth curves with Growthcurver in RGrowthcurver is an R package that analyzes the optical density data by fitting it to a logistic function from which the growth rate, doubling time, and carrying capacity can be calculated. Growthcurver can be installed with the command:
install.packages("growthcurver")
Growthcurver can be run on the formatted tab-delimited file with the following code. The variable "r" contains the growth rate and is extracted once the model has been fit to the data. Note that the units of the growth rate will be based on the units of your file, i.e. if your time is in seconds and the growth measurements are in OD600 the growth rate will be expressed in OD600/second and may need to be converted to other units.
Quality control can be performed by checking for notes on the model fitting. Any samples with low quality fits will be identified by the following code:
Other statistics that can be reported include the carrying capacity (use gc_out$k), the doubling time (gc_out$t_gen), and the inflection point of log phase (gc_out$t_mid) for each sample. | |||||||||
| Changed: | |||||||||
| < < | Additionally, a histogram of the variance of the data can be produced with the following code. Data points should produce a fit similar to a bell curve, with most data points of average variance. Poorly fitting points will appear as outliers on the histogram as in the example below. | ||||||||
| > > | Additionally, a histogram of the variance of the data can be produced with the following code. Poorly fit samples will appear as outliers on the histogram as in the example below. | ||||||||
 | |||||||||
| Added: | |||||||||
| > > | |||||||||
Calculating growth rates using the Grofit R packageGrowth curves can also be fit using Grofit, however this package is no longer supported by the current version of R.References
Contributors
| |||||||||
Revision 132021-11-05 - IsaacGifford
Measuring Microbial Growth Rates in a Plate ReaderThe following protocol can be used to determine the growth rate of a bacterial culture using a plate reader by measuring the optical density (OD600) of the culture over time. This protocol is written specifically for a Tecan Infinite® M200 Pro plate reader combined with the Magellan™ data analysis software and thus may require alteration for other machines. The Nunc™ MicroWell™ 96-Well Optical-Bottom Plates (black wells with clear bottoms) are ideal for this assay.Program set-upThe parameters used by the Barrick Lab are:
Reviving cultures (2 days before experiment)1) Inoculate an overnight culture of each strain being tested. Prepare a separate tube of uninoculated media as a control for contamination. Incubate overnight.Preconditioning cultures (1 day before experiment)Preconditioning acclimates the strains to the media. Additionally, on this day you should pre-warm your media as it takes a long time for the plate reader to warm up media. Not doing this will lead to inconsistent lag time. 2) Inoculate 5 μl of each overnight culture into fresh media to precondition. Prepare a separate tube of uninoculated media as a control for contamination. Incubate overnight.3) Place the media you will use for the assay in an incubator at the correct growth temperature overnight to pre-warm. Growing cells in plate reader and measuring OD600All strains tested should have at least have 3 replicates, although more replicates should be performed as long as there is available space on the plate. Evaporation can occur in the outermost wells, so if there are few enough samples the outer ring of wells should be skipped. Due to small variations in temperature throughout a 96-well plate, best results will be obtained if replicates are distributed randomly across the plate. 4) Begin heating the plate reader to the desired growth temperature using the “Temp. control...” button in the “Start Measurement” dialog box.
6) Inoculate each test well with 5 μl of overnight culture. 7) Place the plate into the plate reader. The lid can be removed (we've had no problems with contamination, as seen on wells with LB blanks, with this). 8) Start your program. 9) Once the program has finished, export the data as an Excel spreadsheet.
Formatting the data | ||||||||
| Changed: | ||||||||
| < < | In order to run Growthcurver, you need to create a tab-delimited file in the following arrangement (see example file): | |||||||
| > > | In order to run Growthcurver, you need to create a tab-delimited file in the following arrangement (see example file below): | |||||||
| ||||||||
| Added: | ||||||||
| > > |
| |||||||
Fitting growth curves with Growthcurver in RGrowthcurver is an R package that analyzes the optical density data by fitting it to a logistic function from which the growth rate, doubling time, and carrying capacity can be calculated. Growthcurver can be installed with the command:
install.packages("growthcurver")
Growthcurver can be run on the formatted tab-delimited file with the following code. The variable "r" contains the growth rate and is extracted once the model has been fit to the data. Note that the units of the growth rate will be based on the units of your file, i.e. if your time is in seconds and the growth measurements are in OD600 the growth rate will be expressed in OD600/second and may need to be converted to other units.
Quality control can be performed by checking for notes on the model fitting. Any samples with low quality fits will be identified by the following code:
Other statistics that can be reported include the carrying capacity (use gc_out$k), the doubling time (gc_out$t_gen), and the inflection point of log phase (gc_out$t_mid) for each sample. | ||||||||
| Changed: | ||||||||
| < < | Additionally, a histogram of the variance of the data can be produced with the following code. Data points should produce a fit similar to a bell curve, with most data points of average variance. Poorly fitting points will appear as outliers on the histogram. | |||||||
| > > | Additionally, a histogram of the variance of the data can be produced with the following code. Data points should produce a fit similar to a bell curve, with most data points of average variance. Poorly fitting points will appear as outliers on the histogram as in the example below. | |||||||
| ||||||||
| Added: | ||||||||
| > > |  | |||||||
Calculating growth rates using the Grofit R packageGrowth curves can also be fit using Grofit, however this package is no longer supported by the current version of R.References
Contributors
| ||||||||
| Added: | ||||||||
| > > |
| |||||||
Revision 122021-11-04 - IsaacGifford
Measuring Microbial Growth Rates in a Plate ReaderThe following protocol can be used to determine the growth rate of a bacterial culture using a plate reader by measuring the optical density (OD600) of the culture over time. This protocol is written specifically for a Tecan Infinite® M200 Pro plate reader combined with the Magellan™ data analysis software and thus may require alteration for other machines. The Nunc™ MicroWell™ 96-Well Optical-Bottom Plates (black wells with clear bottoms) are ideal for this assay.Program set-upThe parameters used by the Barrick Lab are:
Reviving cultures (2 days before experiment)1) Inoculate an overnight culture of each strain being tested. Prepare a separate tube of uninoculated media as a control for contamination. Incubate overnight.Preconditioning cultures (1 day before experiment)Preconditioning acclimates the strains to the media. Additionally, on this day you should pre-warm your media as it takes a long time for the plate reader to warm up media. Not doing this will lead to inconsistent lag time. 2) Inoculate 5 μl of each overnight culture into fresh media to precondition. Prepare a separate tube of uninoculated media as a control for contamination. Incubate overnight.3) Place the media you will use for the assay in an incubator at the correct growth temperature overnight to pre-warm. Growing cells in plate reader and measuring OD600All strains tested should have at least have 3 replicates, although more replicates should be performed as long as there is available space on the plate. Evaporation can occur in the outermost wells, so if there are few enough samples the outer ring of wells should be skipped. Due to small variations in temperature throughout a 96-well plate, best results will be obtained if replicates are distributed randomly across the plate. 4) Begin heating the plate reader to the desired growth temperature using the “Temp. control...” button in the “Start Measurement” dialog box.
6) Inoculate each test well with 5 μl of overnight culture. 7) Place the plate into the plate reader. The lid can be removed (we've had no problems with contamination, as seen on wells with LB blanks, with this). 8) Start your program. 9) Once the program has finished, export the data as an Excel spreadsheet.
| ||||||||
| Changed: | ||||||||
| < < | 10) Average the blank measurements at each time point. Subtract these values from the measurement of each sample at the corresponding time point. | |||||||
| > > | 10) Average the blank measurements at each time point. | |||||||
| Added: | ||||||||
| > > | Formatting the dataIn order to run Growthcurver, you need to create a tab-delimited file in the following arrangement (see example file):
| |||||||
Fitting growth curves with Growthcurver in RGrowthcurver is an R package that analyzes the optical density data by fitting it to a logistic function from which the growth rate, doubling time, and carrying capacity can be calculated. Growthcurver can be installed with the command:
install.packages("growthcurver")
| ||||||||
| Changed: | ||||||||
| < < | Growthcurver can be run with the following code. The input file should be a tab-delimited file, which can be created from the Excel spreadsheet. The variable "r" contains the growth rate and is extracted once the model has been fit to the data. | |||||||
| > > | Growthcurver can be run on the formatted tab-delimited file with the following code. The variable "r" contains the growth rate and is extracted once the model has been fit to the data. Note that the units of the growth rate will be based on the units of your file, i.e. if your time is in seconds and the growth measurements are in OD600 the growth rate will be expressed in OD600/second and may need to be converted to other units. | |||||||
| ||||||||
| Changed: | ||||||||
| < < | Quality control can be performed by checking for notes on the model fitting. Any low quality fits should be identified by the following code: | |||||||
| > > | Quality control can be performed by checking for notes on the model fitting. Any samples with low quality fits will be identified by the following code: | |||||||
| ||||||||
| Added: | ||||||||
| > > | library("dplyr") | |||||||
| gc_out %>% filter(note = "") | ||||||||
| Deleted: | ||||||||
| < < | gc_fit$vals$note | |||||||
| Added: | ||||||||
| > > | Other statistics that can be reported include the carrying capacity (use gc_out$k), the doubling time (gc_out$t_gen), and the inflection point of log phase (gc_out$t_mid) for each sample. | |||||||
Additionally, a histogram of the variance of the data can be produced with the following code. Data points should produce a fit similar to a bell curve, with most data points of average variance. Poorly fitting points will appear as outliers on the histogram.
| ||||||||
| Deleted: | ||||||||
| < < | library(dplyr) | |||||||
| gc_out = as_data_frame(gc_out) hist(gc_out$sigma, main="Histogram of sigma values", xlab = "sigma") | ||||||||
| Changed: | ||||||||
| < < | Calculating Growth Rates using Grofit R package | |||||||
| > > | Calculating growth rates using the Grofit R package | |||||||
Growth curves can also be fit using Grofit, however this package is no longer supported by the current version of R.
References
Contributors
| ||||||||
Revision 112021-11-04 - IsaacGifford
Measuring Microbial Growth Rates in a Plate Reader | ||||||||
| Changed: | ||||||||
| < < | The following protocol can be used to determine the growth rate of a bacterial culture using a plate reader by measuring the optical density (OD600) of the culture over time. This protocol is written specifically for a Tecan Infinite M200 Pro plate reader combined with the Magellan data analysis software and thus may require alteration for other machines. | |||||||
| > > | The following protocol can be used to determine the growth rate of a bacterial culture using a plate reader by measuring the optical density (OD600) of the culture over time. This protocol is written specifically for a Tecan Infinite® M200 Pro plate reader combined with the Magellan™ data analysis software and thus may require alteration for other machines. The Nunc™ MicroWell™ | |||||||
| Added: | ||||||||
| > > | 96-Well Optical-Bottom Plates (black wells with clear bottoms) are ideal for this assay. | |||||||
Program set-upThe parameters used by the Barrick Lab are:
Reviving cultures (2 days before experiment) | ||||||||
| Changed: | ||||||||
| < < | 1) Grow an overnight culture of each strain being tested. Inoculate 2 μl of frozen glycerol stocks of each strain into 5 ml of media. Prepare a separate tube of uninoculated media as a control for contamination. Incubate overnight. | |||||||
| > > | 1) Inoculate an overnight culture of each strain being tested. Prepare a separate tube of uninoculated media as a control for contamination. Incubate overnight. | |||||||
Preconditioning cultures (1 day before experiment)Preconditioning acclimates the strains to the media. Additionally, on this day you should pre-warm your media as it takes a long time for the plate reader to warm up media. Not doing this will lead to inconsistent lag time. 2) Inoculate 5 μl of each overnight culture into fresh media to precondition. Prepare a separate tube of uninoculated media as a control for contamination. Incubate overnight.3) Place the media you will use for the assay in an incubator at the correct growth temperature overnight to pre-warm. Growing cells in plate reader and measuring OD600 | ||||||||
| Changed: | ||||||||
| < < | All strains tested should have at least have 3 replicates, although more replicates should be performed as long as there is available space on the plate. Evaporation can occur in the outermost wells, so if there are few enough samples the outer ring of wells should be skipped. Due to small variations in temperature throughout a 96-well plate, best results will be obtained if replicates are distributed randomly across the plate. Clear Costar 96-well plates are a good brand to use. | |||||||
| > > | All strains tested should have at least have 3 replicates, although more replicates should be performed as long as there is available space on the plate. Evaporation can occur in the outermost wells, so if there are few enough samples the outer ring of wells should be skipped. Due to small variations in temperature throughout a 96-well plate, best results will be obtained if replicates are distributed randomly across the plate. | |||||||
| Changed: | ||||||||
| < < | 4) Add 195 μl of pre-warmed media to each well being used for cultures and additional wells for blanks. 5) Inoculate each test well with 5 μl of overnight culture. 6) Place the plate into the plate reader. The lid can be removed (we've had no problems with contamination, as seen on wells with LB blanks, with this). 7) Start your program. 8) Once the program has finished, export the data as an Excel spreadsheet. | |||||||
| > > | 4) Begin heating the plate reader to the desired growth temperature using the “Temp. control...” button in the “Start Measurement” dialog box.
6) Inoculate each test well with 5 μl of overnight culture. 7) Place the plate into the plate reader. The lid can be removed (we've had no problems with contamination, as seen on wells with LB blanks, with this). | |||||||
| Added: | ||||||||
| > > | 8) Start your program. 9) Once the program has finished, export the data as an Excel spreadsheet.
| |||||||
Fitting growth curves with Growthcurver in RGrowthcurver is an R package that analyzes the optical density data by fitting it to a logistic function from which the growth rate, doubling time, and carrying capacity can be calculated. Growthcurver can be installed with the command:
install.packages("growthcurver")
Growthcurver can be run with the following code. The input file should be a tab-delimited file, which can be created from the Excel spreadsheet. The variable "r" contains the growth rate and is extracted once the model has been fit to the data.
Quality control can be performed by checking for notes on the model fitting. Any low quality fits should be identified by the following code:
Additionally, a histogram of the variance of the data can be produced with the following code. Data points should produce a fit similar to a bell curve, with most data points of average variance. Poorly fitting points will appear as outliers on the histogram.
Calculating Growth Rates using Grofit R packageGrowth curves can also be fit using Grofit, however this package is no longer supported by the current version of R.References
Contributors
| ||||||||
Revision 102021-11-04 - IsaacGifford
Measuring Microbial Growth Rates in a Plate ReaderThe following protocol can be used to determine the growth rate of a bacterial culture using a plate reader by measuring the optical density (OD600) of the culture over time. This protocol is written specifically for a Tecan Infinite M200 Pro plate reader combined with the Magellan data analysis software and thus may require alteration for other machines.Program set-upThe parameters used by the Barrick Lab are:
| ||||||||
| Changed: | ||||||||
| < < | Duration: 16-24 hours (as appropriate for your experiment) Kinetic interval: every 10 minutes Orbital Shaking: 420 seconds at amplitude 3 Wait: 5 seconds Absorbance reading: 600 nm, 25 flashes, 50 ms settle time | |||||||
| > > |
| |||||||
It's important for the program to shake for most of the time that you are not making measurements. Less shaking leads to slower growth.
Reviving cultures (2 days before experiment)1) Grow an overnight culture of each strain being tested. Inoculate 2 μl of frozen glycerol stocks of each strain into 5 ml of media. Prepare a separate tube of uninoculated media as a control for contamination. Incubate overnight.Preconditioning cultures (1 day before experiment)Preconditioning acclimates the strains to the media. Additionally, on this day you should pre-warm your media as it takes a long time for the plate reader to warm up media. Not doing this will lead to inconsistent lag time. 2) Inoculate 5 μl of each overnight culture into fresh media to precondition. Prepare a separate tube of uninoculated media as a control for contamination. Incubate overnight.3) Place the media you will use for the assay in an incubator at the correct growth temperature overnight to pre-warm. Growing cells in plate reader and measuring OD600All strains tested should have at least have 3 replicates, although more replicates should be performed as long as there is available space on the plate. Evaporation can occur in the outermost wells, so if there are few enough samples the outer ring of wells should be skipped. Due to small variations in temperature throughout a 96-well plate, best results will be obtained if replicates are distributed randomly across the plate. Clear Costar 96-well plates are a good brand to use. 4) Add 195 μl of pre-warmed media to each well being used for cultures and additional wells for blanks.5) Inoculate each test well with 5 μl of overnight culture. 6) Place the plate into the plate reader. The lid can be removed (we've had no problems with contamination, as seen on wells with LB blanks, with this). 7) Start your program. 8) Once the program has finished, export the data as an Excel spreadsheet. Fitting growth curves with Growthcurver in RGrowthcurver is an R package that analyzes the optical density data by fitting it to a logistic function from which the growth rate, doubling time, and carrying capacity can be calculated. Growthcurver can be installed with the command:
install.packages("growthcurver")
Growthcurver can be run with the following code. The input file should be a tab-delimited file, which can be created from the Excel spreadsheet. The variable "r" contains the growth rate and is extracted once the model has been fit to the data.
Quality control can be performed by checking for notes on the model fitting. Any low quality fits should be identified by the following code:
Additionally, a histogram of the variance of the data can be produced with the following code. Data points should produce a fit similar to a bell curve, with most data points of average variance. Poorly fitting points will appear as outliers on the histogram.
Calculating Growth Rates using Grofit R packageGrowth curves can also be fit using Grofit, however this package is no longer supported by the current version of R.References
Contributors
| ||||||||
Revision 92021-11-03 - IsaacGifford
Measuring Microbial Growth Rates in a Plate Reader | ||||||||
| Changed: | ||||||||
| < < | The following protocol can be used to determine the growth rate of a bacterial culture using a plate reader by measuring the optical density (OD600) of the culture over time. | |||||||
| > > | The following protocol can be used to determine the growth rate of a bacterial culture using a plate reader by measuring the optical density (OD600) of the culture over time. This protocol is written specifically for a Tecan Infinite M200 Pro plate reader combined with the Magellan data analysis software and thus may require alteration for other machines. | |||||||
Program set-up | ||||||||
| Changed: | ||||||||
| < < | The parameters the Barrick Lab uses are: | |||||||
| > > | The parameters used by the Barrick Lab are: | |||||||
Duration: 16-24 hours (as appropriate for your experiment) Kinetic interval: every 10 minutes Orbital Shaking: 420 seconds at amplitude 3 Wait: 5 seconds Absorbance reading: 600 nm, 25 flashes, 50 ms settle time It's important for the program to shake for most of the time that you are not making measurements. Less shaking leads to slower growth.Reviving cultures (2 days before experiment)1) Grow an overnight culture of each strain being tested. Inoculate 2 μl of frozen glycerol stocks of each strain into 5 ml of media. Prepare a separate tube of uninoculated media as a control for contamination. Incubate overnight.Preconditioning cultures (1 day before experiment)Preconditioning acclimates the strains to the media. Additionally, on this day you should pre-warm your media as it takes a long time for the plate reader to warm up media. Not doing this will lead to inconsistent lag time. | ||||||||
| Changed: | ||||||||
| < < | 2) Inoculate 5 μl of each overnight culture into fresh media to precondition. Prepare a separate tube of uninoculated media as a control for contamination. Incubate overnight. | |||||||
| > > | 2) Inoculate 5 μl of each overnight culture into fresh media to precondition. Prepare a separate tube of uninoculated media as a control for contamination. Incubate overnight. | |||||||
3) Place the media you will use for the assay in an incubator at the correct growth temperature overnight to pre-warm.
Growing cells in plate reader and measuring OD600All strains tested should have at least have 3 replicates, although more replicates should be performed as long as there is available space on the plate. Evaporation can occur in the outermost wells, so if there are few enough samples the outer ring of wells should be skipped. Due to small variations in temperature throughout a 96-well plate, best results will be obtained if replicates are distributed randomly across the plate. Clear Costar 96-well plates are a good brand to use. | ||||||||
| Changed: | ||||||||
| < < | 4) Add 195 μl of pre-warmed media to each well being used for cultures and additional wells for blanks. 5) Inoculate each test well with 5 μl of overnight culture. 6) Place the plate into the plate reader. The lid can be removed (we've had no contamination, as seen on wells with LB blanks, or problems with this) 7) Start your program. 8) Once the program has finished, export the data as an Excel spreadsheet. | |||||||
| > > | 4) Add 195 μl of pre-warmed media to each well being used for cultures and additional wells for blanks. 5) Inoculate each test well with 5 μl of overnight culture. 6) Place the plate into the plate reader. The lid can be removed (we've had no problems with contamination, as seen on wells with LB blanks, with this). 7) Start your program. 8) Once the program has finished, export the data as an Excel spreadsheet. | |||||||
| Changed: | ||||||||
| < < | Fitting growth curves with Growthcurver | |||||||
| > > | Fitting growth curves with Growthcurver in R | |||||||
| Changed: | ||||||||
| < < | Growthcurver analyzes the optical density data by fitting it to a logistic function from which the growth rate, doubling time, and carrying capacity can be calculated. | |||||||
| > > | Growthcurver is an R package that analyzes the optical density data by fitting it to a logistic function from which the growth rate, doubling time, and carrying capacity can be calculated. Growthcurver can be installed with the command: | |||||||
| Added: | ||||||||
| > > |
install.packages("growthcurver")
| |||||||
| Changed: | ||||||||
| < < | Section in progress | |||||||
| > > | Growthcurver can be run with the following code. The input file should be a tab-delimited file, which can be created from the Excel spreadsheet. The variable "r" contains the growth rate and is extracted once the model has been fit to the data. | |||||||
| Added: | ||||||||
| > > |
| |||||||
| Added: | ||||||||
| > > | #import data
growth = read.table("/path/to/your/file/data.tsv", header=T, sep="\t", stringsAsFactors=F)
#calculate growth rate
gc_out = SummarizeGrowthByPlate(growth, bg_correct="blank")
gc_out$r
Quality control can be performed by checking for notes on the model fitting. Any low quality fits should be identified by the following code:
Additionally, a histogram of the variance of the data can be produced with the following code. Data points should produce a fit similar to a bell curve, with most data points of average variance. Poorly fitting points will appear as outliers on the histogram.
Calculating Growth Rates using Grofit R packageGrowth curves can also be fit using Grofit, however this package is no longer supported by the current version of R. | |||||||
References
Contributors
| ||||||||
| Deleted: | ||||||||
| < < | AppendixThe following are instructions for calculating growth rates using Grofit, an R package that is no longer supported by the current version of R.Converting your file with OD data to the proper formatWhat is most critical is the format and file type of the input data file containing the OD600 values. Most plate readers produce an Excel worksheet with the data results. With such a file, the first thing that must be done is to convert it to a comma delimited file. This is simply done by using "save as" in Excel and selecting *.csv as the file format. Your file should look similar to the one provided as example here:
Create a "times only" csv fileGrofit needs an additional file that will only have the times (e.g., 10, 20, 30 mins ...) for every time point measurement. An example of this file is given here:
Calculating Growth Rates using Grofit R packageThe following is a short script (by Julie Perreau) on "R studio" that performs the downloading, installing, running and printing out results. You execute each line in the script by pressing Ctrl+Enter. For each one, you must check no errors are given. You can use the example timesonlyworksheet.csv and yourdatafile.csv files provided above as a test. setwd("C:/.../R_GrowthData") #sets the working directory to the folder (here named R_GrowthData) you'll be working on your computerinstall.packages(c("grofit", "tidyr", "reshape2")) library(grofit) library(tidyr) library(reshape2) growthdata <- read.csv("yourdatafile.csv",sep=",", header=TRUE, check.names = FALSE) <br/> timedata <- read.csv("timesonlyworksheet.csv", sep=",", header=TRUE, check.names = FALSE) <br/> gro <- grofit(timedata, growthdata) #runs grofit <br/> summary_table<-summary.gcFit(gro$gcFit) #makes a summary table <br/> write.csv(summary_table,"FinalTable.csv") #makes a csv file of the summary table Accepting/declining model fits:Once Grofit is run, you will be prompted to answer yes (y) or no (n) to accept or decline the model fits generated for each individual well. Most likely, all you need to do is say yes to all of them, unless there is some obvious model fit mistake which would make some sample unreliable. It is normal for Grofit to be unable to fit the curves to one of the 4 models tried, thus the ERROR sometimes given is not going to affect the final outcome, it just means it couldn't use one of the models to determine the best fit.Reading summary table results:The table summary file produced by Grofit will give you all the parameter values (mu=GrowthRates, lambda=LagPhaseTime and A=MaxAbsorbance) generated from the best model fits. Grofit uses 4 possible models; see documentation for more information.References
| |||||||
| ||||||||
Revision 82021-11-03 - IsaacGifford
Measuring Microbial Growth Rates in a Plate Reader | |||||||||
| Added: | |||||||||
| > > | The following protocol can be used to determine the growth rate of a bacterial culture using a plate reader by measuring the optical density (OD600) of the culture over time.
Program set-upThe parameters the Barrick Lab uses are:
Duration: 16-24 hours (as appropriate for your experiment) Kinetic interval: every 10 minutes Orbital Shaking: 420 seconds at amplitude 3 Wait: 5 seconds Absorbance reading: 600 nm, 25 flashes, 50 ms settle time It's important for the program to shake for most of the time that you are not making measurements. Less shaking leads to slower growth.Reviving cultures (2 days before experiment)1) Grow an overnight culture of each strain being tested. Inoculate 2 μl of frozen glycerol stocks of each strain into 5 ml of media. Prepare a separate tube of uninoculated media as a control for contamination. Incubate overnight.Preconditioning cultures (1 day before experiment)Preconditioning acclimates the strains to the media. Additionally, on this day you should pre-warm your media as it takes a long time for the plate reader to warm up media. Not doing this will lead to inconsistent lag time. 2) Inoculate 5 μl of each overnight culture into fresh media to precondition. Prepare a separate tube of uninoculated media as a control for contamination. Incubate overnight. 3) Place the media you will use for the assay in an incubator at the correct growth temperature overnight to pre-warm. | ||||||||
Growing cells in plate reader and measuring OD600 | |||||||||
| Changed: | |||||||||
| < < | All strains tested should at least have 3 replicates. Due to small variations in temperature throughout a 96-well plate, best results will be obtained if replicates are randomly located at different wells. Set up 200 µL cultures with 2 µL from a pre-conditioned (same media as actual test) overnight culture. These 100-fold dilutions should be good enough for OD600 measurements that will give out reliable information for lag phase, exponential phase and stationary phase. To allow cultures to aerate properly, set the plate reader to shake (1 mm radius) at least every 5 minutes; normally this would be before and after taking reads. | ||||||||
| > > | All strains tested should have at least have 3 replicates, although more replicates should be performed as long as there is available space on the plate. Evaporation can occur in the outermost wells, so if there are few enough samples the outer ring of wells should be skipped. Due to small variations in temperature throughout a 96-well plate, best results will be obtained if replicates are distributed randomly across the plate. Clear Costar 96-well plates are a good brand to use. | ||||||||
| Changed: | |||||||||
| < < | Other advice:
| ||||||||
| > > | 4) Add 195 μl of pre-warmed media to each well being used for cultures and additional wells for blanks. 5) Inoculate each test well with 5 μl of overnight culture. 6) Place the plate into the plate reader. The lid can be removed (we've had no contamination, as seen on wells with LB blanks, or problems with this) | ||||||||
| Added: | |||||||||
| > > | 7) Start your program. 8) Once the program has finished, export the data as an Excel spreadsheet. | ||||||||
| Changed: | |||||||||
| < < | Good 96-well plates to use clear Costar brand. Make sure you remove the lid before inserting in the plate reader, we've had no contamination (as seen on wells with LB blanks) or problems with this. | ||||||||
| > > | Fitting growth curves with Growthcurver | ||||||||
| Added: | |||||||||
| > > | Growthcurver analyzes the optical density data by fitting it to a logistic function from which the growth rate, doubling time, and carrying capacity can be calculated.
Section in progress
References
Contributors
AppendixThe following are instructions for calculating growth rates using Grofit, an R package that is no longer supported by the current version of R. | ||||||||
Converting your file with OD data to the proper formatWhat is most critical is the format and file type of the input data file containing the OD600 values. Most plate readers produce an Excel worksheet with the data results. With such a file, the first thing that must be done is to convert it to a comma delimited file. This is simply done by using "save as" in Excel and selecting *.csv as the file format. Your file should look similar to the one provided as example here:
Create a "times only" csv fileGrofit needs an additional file that will only have the times (e.g., 10, 20, 30 mins ...) for every time point measurement. An example of this file is given here:
Calculating Growth Rates using Grofit R packageThe following is a short script (by Julie Perreau) on "R studio" that performs the downloading, installing, running and printing out results. You execute each line in the script by pressing Ctrl+Enter. For each one, you must check no errors are given. You can use the example timesonlyworksheet.csv and yourdatafile.csv files provided above as a test. setwd("C:/.../R_GrowthData") #sets the working directory to the folder (here named R_GrowthData) you'll be working on your computerinstall.packages(c("grofit", "tidyr", "reshape2")) library(grofit) library(tidyr) library(reshape2) growthdata <- read.csv("yourdatafile.csv",sep=",", header=TRUE, check.names = FALSE) <br/> timedata <- read.csv("timesonlyworksheet.csv", sep=",", header=TRUE, check.names = FALSE) <br/> gro <- grofit(timedata, growthdata) #runs grofit <br/> summary_table<-summary.gcFit(gro$gcFit) #makes a summary table <br/> write.csv(summary_table,"FinalTable.csv") #makes a csv file of the summary table Accepting/declining model fits:Once Grofit is run, you will be prompted to answer yes (y) or no (n) to accept or decline the model fits generated for each individual well. Most likely, all you need to do is say yes to all of them, unless there is some obvious model fit mistake which would make some sample unreliable. It is normal for Grofit to be unable to fit the curves to one of the 4 models tried, thus the ERROR sometimes given is not going to affect the final outcome, it just means it couldn't use one of the models to determine the best fit.Reading summary table results:The table summary file produced by Grofit will give you all the parameter values (mu=GrowthRates, lambda=LagPhaseTime and A=MaxAbsorbance) generated from the best model fits. Grofit uses 4 possible models; see documentation for more information.References | |||||||||
| Deleted: | |||||||||
| < < |
Contributors
| ||||||||
| |||||||||
Revision 72021-06-03 - JeffreyBarrick
| |||||||||
| Changed: | |||||||||
| < < | Growth Rates using "R studio" | ||||||||
| > > | Measuring Microbial Growth Rates in a Plate Reader | ||||||||
| Deleted: | |||||||||
| < < | Determining growth rates for many cultures or from plate reader data can be a daunting task. To automate this process, we've found that the Grofit package for R can be very useful and reliable.
Download and install "R" and "R studio"You'll need to download and install R on your computer. Once R is installed, you can then download and install "R studio" which is easier to work with. You can download "R" here: https://cran.r-project.org/mirrors.htmlYou can download "R studio" here: https://www.rstudio.com/ | ||||||||
Growing cells in plate reader and measuring OD600All strains tested should at least have 3 replicates. Due to small variations in temperature throughout a 96-well plate, best results will be obtained if replicates are randomly located at different wells. Set up 200 µL cultures with 2 µL from a pre-conditioned (same media as actual test) overnight culture. These 100-fold dilutions should be good enough for OD600 measurements that will give out reliable information for lag phase, exponential phase and stationary phase. To allow cultures to aerate properly, set the plate reader to shake (1 mm radius) at least every 5 minutes; normally this would be before and after taking reads. | |||||||||
| Added: | |||||||||
| > > |
Other advice:
| ||||||||
Good 96-well plates to use clear Costar brand. Make sure you remove the lid before inserting in the plate reader, we've had no contamination (as seen on wells with LB blanks) or problems with this.
Converting your file with OD data to the proper formatWhat is most critical is the format and file type of the input data file containing the OD600 values. Most plate readers produce an Excel worksheet with the data results. With such a file, the first thing that must be done is to convert it to a comma delimited file. This is simply done by using "save as" in Excel and selecting *.csv as the file format. Your file should look similar to the one provided as example here:
Create a "times only" csv fileGrofit needs an additional file that will only have the times (e.g., 10, 20, 30 mins ...) for every time point measurement. An example of this file is given here:
Calculating Growth Rates using Grofit R packageThe following is a short script (by Julie Perreau) on "R studio" that performs the downloading, installing, running and printing out results. You execute each line in the script by pressing Ctrl+Enter. For each one, you must check no errors are given. You can use the example timesonlyworksheet.csv and yourdatafile.csv files provided above as a test. setwd("C:/.../R_GrowthData") #sets the working directory to the folder (here named R_GrowthData) you'll be working on your computerinstall.packages(c("grofit", "tidyr", "reshape2")) library(grofit) library(tidyr) library(reshape2) growthdata <- read.csv("yourdatafile.csv",sep=",", header=TRUE, check.names = FALSE) <br/> timedata <- read.csv("timesonlyworksheet.csv", sep=",", header=TRUE, check.names = FALSE) <br/> gro <- grofit(timedata, growthdata) #runs grofit <br/> summary_table<-summary.gcFit(gro$gcFit) #makes a summary table <br/> write.csv(summary_table,"FinalTable.csv") #makes a csv file of the summary table Accepting/declining model fits:Once Grofit is run, you will be prompted to answer yes (y) or no (n) to accept or decline the model fits generated for each individual well. Most likely, all you need to do is say yes to all of them, unless there is some obvious model fit mistake which would make some sample unreliable. It is normal for Grofit to be unable to fit the curves to one of the 4 models tried, thus the ERROR sometimes given is not going to affect the final outcome, it just means it couldn't use one of the models to determine the best fit.Reading summary table results:The table summary file produced by Grofit will give you all the parameter values (mu=GrowthRates, lambda=LagPhaseTime and A=MaxAbsorbance) generated from the best model fits. Grofit uses 4 possible models; see documentation for more information.References
Contributors
| |||||||||
Revision 62017-11-06 - GabrielSuarez
| |||||||||
| Deleted: | |||||||||
| < < | |||||||||
| Changed: | |||||||||
| < < | Determining growth rates for many cultures or from plate reader data can be a daunting task. To automate this process, we've found that the Grofit package for R can be very useful and reliable. | ||||||||
| > > | Growth Rates using "R studio" | ||||||||
| Added: | |||||||||
| > > | Determining growth rates for many cultures or from plate reader data can be a daunting task. To automate this process, we've found that the Grofit package for R can be very useful and reliable. | ||||||||
Download and install "R" and "R studio" | |||||||||
| Changed: | |||||||||
| < < | You'll need to download and install R on your computer. Once R is installed, you can then download and install "R studio" which is easier to work with. | ||||||||
| > > | You'll need to download and install R on your computer. Once R is installed, you can then download and install "R studio" which is easier to work with. | ||||||||
| Changed: | |||||||||
| < < | You can download "R" here: https://cran.r-project.org/mirrors.html | ||||||||
| > > | You can download "R" here: https://cran.r-project.org/mirrors.html You can download "R studio" here: https://www.rstudio.com/ | ||||||||
| Deleted: | |||||||||
| < < | You can download "R studio" here: https://www.rstudio.com/ | ||||||||
Growing cells in plate reader and measuring OD600 | |||||||||
| Changed: | |||||||||
| < < | All strains tested should at least have at least 3 replicates. Due to small variations in temperature throughout a 96-well plate, best results will be obtained if replicates are randomly located at different wells. | ||||||||
| > > | All strains tested should at least have 3 replicates. Due to small variations in temperature throughout a 96-well plate, best results will be obtained if replicates are randomly located at different wells. Set up 200 µL cultures with 2 µL from a pre-conditioned (same media as actual test) overnight culture. These 100-fold dilutions should be good enough for OD600 measurements that will give out reliable information for lag phase, exponential phase and stationary phase. To allow cultures to aerate properly, set the plate reader to shake (1 mm radius) at least every 5 minutes; normally this would be before and after taking reads. | ||||||||
| Deleted: | |||||||||
| < < | Set up 200 µL cultures with 2 µL from a pre-conditioned (same media as actual test) overnight culture. These 100-fold dilutions should be good enough for OD600 measurements that will give out reliable information for lag phase, exponential phase and stationary phase. To allow cultures to aerate properly, set the plate reader to shake (1 mm radius) at least every 5 minutes; normally this would be before and after taking reads. | ||||||||
| Changed: | |||||||||
| < < | Good 96-well plates to use clear Costar brand. Make sure you remove the lid before inserting in the plate reader, we've had no contamination (as seen on wells with LB blanks) or problems with this. | ||||||||
| > > | Good 96-well plates to use clear Costar brand. Make sure you remove the lid before inserting in the plate reader, we've had no contamination (as seen on wells with LB blanks) or problems with this. | ||||||||
Converting your file with OD data to the proper format | |||||||||
| Changed: | |||||||||
| < < | What is most critical is the format and file type of the input data file containing the OD600 values. Most plate readers produce an Excel worksheet with the data results. With such a file, the first thing that must be done is to convert it to a comma delimited file. This is simply done by using "save as" in Excel and selecting *.csv as the file format. | ||||||||
| > > | What is most critical is the format and file type of the input data file containing the OD600 values. Most plate readers produce an Excel worksheet with the data results. With such a file, the first thing that must be done is to convert it to a comma delimited file. This is simply done by using "save as" in Excel and selecting *.csv as the file format. Your file should look similar to the one provided as example here: | ||||||||
| Deleted: | |||||||||
| < < | Your file should look similar to the one provided as example here: | ||||||||
Create a "times only" csv file | |||||||||
| Changed: | |||||||||
| < < | Grofit needs an additional file that will only have the times (e.g., 10, 20, 30 mins ...) for every time point measurement. | ||||||||
| > > | Grofit needs an additional file that will only have the times (e.g., 10, 20, 30 mins ...) for every time point measurement. An example of this file is given here: | ||||||||
| Deleted: | |||||||||
| < < | An example of this file is given here: | ||||||||
| |||||||||
| Changed: | |||||||||
| < < | Calculating Growth Rates using Grofit R packageThe following is a short script (by Julie Perreau) on "R studio" that performs the downloading, installing, running and printing out results. You execute each line in the script by pressing Ctrl+Enter. For each one, you must check no errors are given. You can use the example timesonlyworksheet.csv and yourdatafile.csv files provided above as a test. | ||||||||
| > > | Calculating Growth Rates using Grofit R packageThe following is a short script (by Julie Perreau) on "R studio" that performs the downloading, installing, running and printing out results. You execute each line in the script by pressing Ctrl+Enter. For each one, you must check no errors are given. You can use the example timesonlyworksheet.csv and yourdatafile.csv files provided above as a test. | ||||||||
| Changed: | |||||||||
| < < | setwd("C:/.../R_GrowthData") #sets the working directory to the folder (here named R_GrowthData) you'll be working on your computer | ||||||||
| > > | setwd("C:/.../R_GrowthData") #sets the working directory to the folder (here named R_GrowthData) you'll be working on your computer install.packages(c("grofit", "tidyr", "reshape2")) library(grofit) library(tidyr) library(reshape2) growthdata <- read.csv("yourdatafile.csv",sep=",", header=TRUE, check.names = FALSE) <br/> timedata <- read.csv("timesonlyworksheet.csv", sep=",", header=TRUE, check.names = FALSE) <br/> gro <- grofit(timedata, growthdata) #runs grofit <br/> summary_table<-summary.gcFit(gro$gcFit) #makes a summary table <br/> write.csv(summary_table,"FinalTable.csv") #makes a csv file of the summary table | ||||||||
| Deleted: | |||||||||
| < < | install.packages(c("grofit", "tidyr", "reshape2")) library(grofit) library(tidyr) library(reshape2) growthdata <- read.csv("yourdatafile.csv",sep=",", header=TRUE, check.names = FALSE) timedata <- read.csv("timesonlyworksheet.csv", sep=",", header=TRUE, check.names = FALSE) gro <- grofit(timedata, growthdata) #runs grofit summary_table<-summary.gcFit(gro$gcFit) #makes a summary table write.csv(summary_table,"FinalTable.csv") #makes a csv file of the summary table | ||||||||
Accepting/declining model fits: | |||||||||
| Changed: | |||||||||
| < < | Once Grofit is run, you will be prompted to answer yes (y) or no (n) to accept or decline the model fits generated for each individual well. Most likely, all you need to do is say yes to all of them, unless there is some obvious model fit mistake which would make some sample unreliable. It is normal for Grofit to be unable to fit the curves to one of the 4 models tried, thus the ERROR sometimes given is not going to affect the final outcome, it just means it couldn't use one of the models to determine the best fit. | ||||||||
| > > | Once Grofit is run, you will be prompted to answer yes (y) or no (n) to accept or decline the model fits generated for each individual well. Most likely, all you need to do is say yes to all of them, unless there is some obvious model fit mistake which would make some sample unreliable. It is normal for Grofit to be unable to fit the curves to one of the 4 models tried, thus the ERROR sometimes given is not going to affect the final outcome, it just means it couldn't use one of the models to determine the best fit. | ||||||||
| Deleted: | |||||||||
| < < | |||||||||
Reading summary table results: | |||||||||
| Changed: | |||||||||
| < < | The table summary file produced by Grofit will give you all the parameter values (mu=GrowthRates, lambda=LagPhaseTime and A=MaxAbsorbance) generated from the best model fits. Grofit uses 4 possible models; see documentation for more information. | ||||||||
| > > | The table summary file produced by Grofit will give you all the parameter values (mu=GrowthRates, lambda=LagPhaseTime and A=MaxAbsorbance) generated from the best model fits. Grofit uses 4 possible models; see documentation for more information. | ||||||||
| Deleted: | |||||||||
| < < | |||||||||
References
Contributors
| |||||||||
| Deleted: | |||||||||
| < < | |||||||||
| |||||||||
Revision 52016-12-16 - JeffreyBarrick
Growth Rates using "R studio"
Determining growth rates for many cultures or from plate reader data can be a daunting task. To automate this process, we've found that the Grofit package for R can be very useful and reliable.
| |||||||||
| Changed: | |||||||||
| < < | All strains tested should at least have 3 replicates. Due to small variations in temperature throughout a 96-well plate, best results will be obtained if replicates are randomly located at different wells. Set up 200µL cultures with 2µL from a pre-conditioned (same media as actual test) overnight culture. These 100-fold dilutions should be good enough for OD600 measurements that will give out reliable information for lag phase, exponential phase and stationary phase. To allow cultures to aerate properly, set the plate reader to shake (1mm radious) at least every 5 minutes; normally this would be before and after taking reads. | ||||||||
| > > | All strains tested should at least have at least 3 replicates. Due to small variations in temperature throughout a 96-well plate, best results will be obtained if replicates are randomly located at different wells. Set up 200 µL cultures with 2 µL from a pre-conditioned (same media as actual test) overnight culture. These 100-fold dilutions should be good enough for OD600 measurements that will give out reliable information for lag phase, exponential phase and stationary phase. To allow cultures to aerate properly, set the plate reader to shake (1 mm radius) at least every 5 minutes; normally this would be before and after taking reads. | ||||||||
| Changed: | |||||||||
| < < | A good 96-well plates to are clear Costar brand. Make sure you remove the lid before inserting in the plate reader, we've had no contamination (as seen on wells with LB blanks) or problems with this. | ||||||||
| > > | Good 96-well plates to use clear Costar brand. Make sure you remove the lid before inserting in the plate reader, we've had no contamination (as seen on wells with LB blanks) or problems with this. | ||||||||
Converting your file with OD data to the proper formatWhat is most critical is the format and file type of the input data file containing the OD600 values. Most plate readers produce an Excel worksheet with the data results. With such a file, the first thing that must be done is to convert it to a comma delimited file. This is simply done by using "save as" in Excel and selecting *.csv as the file format. Your file should look similar to the one provided as example here:
Create a "times only" csv fileGrofit needs an additional file that will only have the times (e.g., 10, 20, 30 mins ...) for every time point measurement. An example of this file is given here:
Calculating Growth Rates using Grofit R packageThe following is a short script (by Julie Perreau) on "R studio" that performs the downloading, installing, running and printing out results. You execute each line in the script by pressing Ctrl+Enter. For each one, you must check no errors are given. You can use the example timesonlyworksheet.csv and yourdatafile.csv files provided above as a test. setwd("C:/.../R_GrowthData") #sets the working directory to the folder (here named R_GrowthData) you'll be working on your computerinstall.packages(c("grofit", "tidyr", "reshape2")) library(grofit) library(tidyr) library(reshape2) growthdata <- read.csv("yourdatafile.csv",sep=",", header=TRUE, check.names = FALSE) timedata <- read.csv("timesonlyworksheet.csv", sep=",", header=TRUE, check.names = FALSE) gro <- grofit(timedata, growthdata) #runs grofit summary_table<-summary.gcFit(gro$gcFit) #makes a summary table write.csv(summary_table,"FinalTable.csv") #makes a csv file of the summary table Accepting/declining model fits:Once Grofit is run, you will be prompted to answer yes (y) or no (n) to accept or decline the model fits generated for each individual well. Most likely, all you need to do is say yes to all of them, unless there is some obvious model fit mistake which would make some sample unreliable. It is normal for Grofit to be unable to fit the curves to one of the 4 models tried, thus the ERROR sometimes given is not going to affect the final outcome, it just means it couldn't use one of the models to determine the best fit.Reading summary table results:The table summary file produced by Grofit will give you all the parameter values (mu=GrowthRates, lambda=LagPhaseTime and A=MaxAbsorbance) generated from the best model fits. Grofit uses 4 possible models; see documentation for more information.References
Contributors
| |||||||||
Revision 42016-12-16 - JeffreyBarrick
Growth Rates using "R studio"
Determining growth rates for many cultures or from plate reader data can be a daunting task. To automate this process, we've found that the Grofit package for R can be very useful and reliable.
| ||||||||
| Changed: | ||||||||
| < < | -- Main.GabrielSuarez - 15 Dec 2016 | |||||||
| > > | ||||||||
| ||||||||
| Changed: | ||||||||
| < < |
| |||||||
| > > |
| |||||||
Revision 32016-12-15 - GabrielSuarez
| |||||||||
| Changed: | |||||||||
| < < | Growth Rates using "R studio" - Overview | ||||||||
| > > | Growth Rates using "R studio" | ||||||||
Determining growth rates for many cultures or from plate reader data can be a daunting task. To automate this process, we've found that the Grofit package for R can be very useful and reliable.
Download and install "R" and "R studio"You'll need to download and install R on your computer. Once R is installed, you can then download and install "R studio" which is easier to work with. You can download "R" here: https://cran.r-project.org/mirrors.htmlYou can download "R studio" here: https://www.rstudio.com/ Growing cells in plate reader and measuring OD600All strains tested should at least have 3 replicates. Due to small variations in temperature throughout a 96-well plate, best results will be obtained if replicates are randomly located at different wells. | |||||||||
| Changed: | |||||||||
| < < | Set up 200µL cultures with 2µL from a pre-conditioned (same media as actual test) overnight culture. These 100-fold dilutions should be good enough for OD600 measurements that will give out reliable information for lag phase, exponential phase and stationary phase. To allow cultures to aerate properly, set the plate reader to shake (1mm radious) at least every 5 minutes; normally this would be before and after taking reads. | ||||||||
| > > | Set up 200µL cultures with 2µL from a pre-conditioned (same media as actual test) overnight culture. These 100-fold dilutions should be good enough for OD600 measurements that will give out reliable information for lag phase, exponential phase and stationary phase. To allow cultures to aerate properly, set the plate reader to shake (1mm radious) at least every 5 minutes; normally this would be before and after taking reads. | ||||||||
| Added: | |||||||||
| > > | A good 96-well plates to are clear Costar brand. Make sure you remove the lid before inserting in the plate reader, we've had no contamination (as seen on wells with LB blanks) or problems with this. | ||||||||
Converting your file with OD data to the proper format | |||||||||
| Changed: | |||||||||
| < < | What is most critical is the format and file type of the input data file containing the OD600 values. Most plate readers produce an Excel worksheet with the data results. With such a file, the first thing that must be done is to convert it to a comma delimited file. This is simply done by using "save as" in Excel and selecting *.csv as the file format. | ||||||||
| > > | What is most critical is the format and file type of the input data file containing the OD600 values. Most plate readers produce an Excel worksheet with the data results. With such a file, the first thing that must be done is to convert it to a comma delimited file. This is simply done by using "save as" in Excel and selecting *.csv as the file format. | ||||||||
| Added: | |||||||||
| > > | Your file should look similar to the one provided as example here:
| ||||||||
Create a "times only" csv fileGrofit needs an additional file that will only have the times (e.g., 10, 20, 30 mins ...) for every time point measurement. An example of this file is given here: | |||||||||
| Added: | |||||||||
| > > |
| ||||||||
| Changed: | |||||||||
| < < | Calculating Mutation Rates using Grofit R packageAs a reference, the following is a short script on "R studio" that performs the downloading, installing, running and printing out results. You execute each line in the script by pressing Ctrl+Enter. For each one, you must check no errors are given. | ||||||||
| > > | Calculating Growth Rates using Grofit R packageThe following is a short script (by Julie Perreau) on "R studio" that performs the downloading, installing, running and printing out results. You execute each line in the script by pressing Ctrl+Enter. For each one, you must check no errors are given. You can use the example timesonlyworksheet.csv and yourdatafile.csv files provided above as a test. | ||||||||
| Changed: | |||||||||
| < < | setwd("C:/.../R_GrowthData") #sets the working directory to the folder (here named R_GrowthData) you'll be working on your computer | ||||||||
| > > | setwd("C:/.../R_GrowthData") #sets the working directory to the folder (here named R_GrowthData) you'll be working on your computer | ||||||||
| install.packages(c("grofit", "tidyr", "reshape2")) library(grofit) library(tidyr) library(reshape2) growthdata <- read.csv("yourdatafile.csv",sep=",", header=TRUE, check.names = FALSE) timedata <- read.csv("timesonlyworksheet.csv", sep=",", header=TRUE, check.names = FALSE) | |||||||||
| Changed: | |||||||||
| < < | gro <- grofit(timedata, growthdata) #runs grofit | ||||||||
| > > | gro <- grofit(timedata, growthdata) #runs grofit | ||||||||
| Changed: | |||||||||
| < < | summary_table<-summary.gcFit(gro$gcFit) #makes a summary table | ||||||||
| > > | summary_table<-summary.gcFit(gro$gcFit) #makes a summary table | ||||||||
write.csv(summary_table,"FinalTable.csv") #makes a csv file of the summary table Accepting/declining model fits: | |||||||||
| Changed: | |||||||||
| < < | Once Grofit is run, you will be prompted to answer yes (y) or no (n) to accept or decline the model fits generated for each individual well. Most likely, all you need to do is say yes to all of them, unless there is some obvious model fit mistake which would make some sample unreliable. | ||||||||
| > > | Once Grofit is run, you will be prompted to answer yes (y) or no (n) to accept or decline the model fits generated for each individual well. Most likely, all you need to do is say yes to all of them, unless there is some obvious model fit mistake which would make some sample unreliable. It is normal for Grofit to be unable to fit the curves to one of the 4 models tried, thus the ERROR sometimes given is not going to affect the final outcome, it just means it couldn't use one of the models to determine the best fit. | ||||||||
| Added: | |||||||||
| > > | |||||||||
Reading summary table results:The table summary file produced by Grofit will give you all the parameter values (mu=GrowthRates, lambda=LagPhaseTime and A=MaxAbsorbance) generated from the best model fits. Grofit uses 4 possible models; see documentation for more information.References | |||||||||
| Added: | |||||||||
| > > | Contributors | ||||||||
| Added: | |||||||||
| > > |
| ||||||||
| -- Main.GabrielSuarez - 15 Dec 2016 | |||||||||
| Added: | |||||||||
| > > |
| ||||||||
Revision 22016-12-15 - JuliePerreau
Growth Rates using "R studio" - OverviewDetermining growth rates for many cultures or from plate reader data can be a daunting task. To automate this process, we've found that the Grofit package for R can be very useful and reliable.Download and install "R" and "R studio"You'll need to download and install R on your computer. Once R is installed, you can then download and install "R studio" which is easier to work with. | ||||||||
| Changed: | ||||||||
| < < | You can download "R" here: https://cran.r-project.org/mirrors.html | |||||||
| > > | You can download "R" here: https://cran.r-project.org/mirrors.html | |||||||
| Deleted: | ||||||||
| < < | ||||||||
| You can download "R studio" here: https://www.rstudio.com/ | ||||||||
| Deleted: | ||||||||
| < < | ||||||||
Growing cells in plate reader and measuring OD600All strains tested should at least have 3 replicates. Due to small variations in temperature throughout a 96-well plate, best results will be obtained if replicates are randomly located at different wells. Set up 200µL cultures with 2µL from a pre-conditioned (same media as actual test) overnight culture. These 100-fold dilutions should be good enough for OD600 measurements that will give out reliable information for lag phase, exponential phase and stationary phase. To allow cultures to aerate properly, set the plate reader to shake (1mm radious) at least every 5 minutes; normally this would be before and after taking reads.Converting your file with OD data to the proper formatWhat is most critical is the format and file type of the input data file containing the OD600 values. Most plate readers produce an Excel worksheet with the data results. With such a file, the first thing that must be done is to convert it to a comma delimited file. This is simply done by using "save as" in Excel and selecting *.csv as the file format.Create a "times only" csv fileGrofit needs an additional file that will only have the times (e.g., 10, 20, 30 mins ...) for every time point measurement. An example of this file is given here:Calculating Mutation Rates using Grofit R packageAs a reference, the following is a short script on "R studio" that performs the downloading, installing, running and printing out results. You execute each line in the script by pressing Ctrl+Enter. For each one, you must check no errors are given. setwd("C:/.../R_GrowthData") #sets the working directory to the folder (here named R_GrowthData) you'll be working on your computer | ||||||||
| Changed: | ||||||||
| < < | install.packages("grofit") library(grofit) install.packages("tidyr") library(tidyr) | |||||||
| > > | install.packages(c("grofit", "tidyr", "reshape2")) library(grofit) library(tidyr) library(reshape2) | |||||||
| Deleted: | ||||||||
| < < | install.packages("reshape2") library(reshape2) | |||||||
| Changed: | ||||||||
| < < | growthdata<-read.csv("yourdatafile.csv",sep=",",header=TRUE,check.names = FALSE) timedata<-read.csv("timesonlyworksheet.csv",sep=",",header=TRUE,check.names = FALSE) gro<-grofit(timedata, growthdata) #runs grofit | |||||||
| > > | growthdata <- read.csv("yourdatafile.csv",sep=",", header=TRUE, check.names = FALSE) timedata <- read.csv("timesonlyworksheet.csv", sep=",", header=TRUE, check.names = FALSE) gro <- grofit(timedata, growthdata) #runs grofit | |||||||
summary_table<-summary.gcFit(gro$gcFit) #makes a summary table write.csv(summary_table,"FinalTable.csv") #makes a csv file of the summary table Accepting/declining model fits:Once Grofit is run, you will be prompted to answer yes (y) or no (n) to accept or decline the model fits generated for each individual well. Most likely, all you need to do is say yes to all of them, unless there is some obvious model fit mistake which would make some sample unreliable.Reading summary table results:The table summary file produced by Grofit will give you all the parameter values (mu=GrowthRates, lambda=LagPhaseTime and A=MaxAbsorbance) generated from the best model fits. Grofit uses 4 possible models; see documentation for more information.References-- Main.GabrielSuarez - 15 Dec 2016 | ||||||||
Revision 12016-12-15 - GabrielSuarez
Growth Rates using "R studio" - OverviewDetermining growth rates for many cultures or from plate reader data can be a daunting task. To automate this process, we've found that the Grofit package for R can be very useful and reliable.Download and install "R" and "R studio"You'll need to download and install R on your computer. Once R is installed, you can then download and install "R studio" which is easier to work with. You can download "R" here: https://cran.r-project.org/mirrors.html You can download "R studio" here: https://www.rstudio.com/Growing cells in plate reader and measuring OD600All strains tested should at least have 3 replicates. Due to small variations in temperature throughout a 96-well plate, best results will be obtained if replicates are randomly located at different wells. Set up 200µL cultures with 2µL from a pre-conditioned (same media as actual test) overnight culture. These 100-fold dilutions should be good enough for OD600 measurements that will give out reliable information for lag phase, exponential phase and stationary phase. To allow cultures to aerate properly, set the plate reader to shake (1mm radious) at least every 5 minutes; normally this would be before and after taking reads.Converting your file with OD data to the proper formatWhat is most critical is the format and file type of the input data file containing the OD600 values. Most plate readers produce an Excel worksheet with the data results. With such a file, the first thing that must be done is to convert it to a comma delimited file. This is simply done by using "save as" in Excel and selecting *.csv as the file format.Create a "times only" csv fileGrofit needs an additional file that will only have the times (e.g., 10, 20, 30 mins ...) for every time point measurement. An example of this file is given here:Calculating Mutation Rates using Grofit R packageAs a reference, the following is a short script on "R studio" that performs the downloading, installing, running and printing out results. You execute each line in the script by pressing Ctrl+Enter. For each one, you must check no errors are given. setwd("C:/.../R_GrowthData") #sets the working directory to the folder (here named R_GrowthData) you'll be working on your computerinstall.packages("grofit") library(grofit) install.packages("tidyr") library(tidyr) install.packages("reshape2") library(reshape2) growthdata<-read.csv("yourdatafile.csv",sep=",",header=TRUE,check.names = FALSE) timedata<-read.csv("timesonlyworksheet.csv",sep=",",header=TRUE,check.names = FALSE) gro<-grofit(timedata, growthdata) #runs grofit summary_table<-summary.gcFit(gro$gcFit) #makes a summary table write.csv(summary_table,"FinalTable.csv") #makes a csv file of the summary table Accepting/declining model fits:Once Grofit is run, you will be prompted to answer yes (y) or no (n) to accept or decline the model fits generated for each individual well. Most likely, all you need to do is say yes to all of them, unless there is some obvious model fit mistake which would make some sample unreliable.Reading summary table results:The table summary file produced by Grofit will give you all the parameter values (mu=GrowthRates, lambda=LagPhaseTime and A=MaxAbsorbance) generated from the best model fits. Grofit uses 4 possible models; see documentation for more information.References-- Main.GabrielSuarez - 15 Dec 2016 |
View topic | History: r15 < r14 < r13 < r12 | More topic actions...
 - Overview
- Overview